13.4. Student’s t-Distribution#
Further Reading: §5.3 in Navidi (2015)
13.4.1. Learning Objectives#
After studying this notebook, attending class, completing the home activities, and asking questions, you should be able to:
Calculate any size confidence interval (95%, 99%, etc.) using z- or t-distribution
Explain why the t-distribution is important. Relate this back to the CLT
Check for the assumption required to apply the t-distribution
# load libraries
import scipy.stats as stats
import numpy as np
import math
import matplotlib.pyplot as plt
13.4.2. Main Idea#
What happens if the sample size is small and we do not know \(\sigma\)? Two complications arise:
Cannot invoke central limit theorem
Need to account for errors when estimating \(\sigma\) (population standard deviation) using \(s\) (sample standard deviation)
Solution: Student’s t-distribution
History Side Tangent
Published in 1908 by William Sealy Goshet under the pseudonym Student.


13.4.3. Degrees of Freedom and Visualization#
\(\nu\) is the degrees of freedom:
Student’s t-distribution converges to the Gaussian distribution (normal distribution shown in last notebook) as \(\nu \rightarrow \infty\)
\(\nu\) is often \(n\) - 1. We lose 1 degree of freedom to estimate standard deviation


13.4.4. Comparison of \(z^*\) and \(t^*\)#
n = 5
print("Consider n =",n)
# calculate zstar
# argument 1: confidence level
zstar95 = stats.norm.interval(0.95)
print("\nz-star for 95% interval:",zstar95)
# calculate tstar
# argument 1: confidence level
# argument 2: degrees of freedom
tstar95 = stats.t.interval(0.95,n-1)
print("\nt-star for 95% interval:",tstar95)
# percent error
print("\nPercent error using z instead of t:",abs((1 - zstar95[0]/tstar95[0]))*100,"%")
Consider n = 5
z-star for 95% interval: (-1.959963984540054, 1.959963984540054)
t-star for 95% interval: (-2.7764451051977987, 2.7764451051977987)
Percent error using z instead of t: 29.407428914377086 %
13.4.5. Catalyst Example Revisited#
Note
Class Discussion: Below is the code from the Confidence Intervals notebook. How would we modify our code to calculate a t-interval (instead of a z-interval)?
The notebook with the original Catalyst Example can be found here.
# catalyst example data
lifetime = [3.2, 6.8, 4.2, 9.2, 11.2, 3.7, 2.9, 12.6, 6.4, 7.5, 8.6,
4.5, 3.0, 9.6, 1.5, 4.5, 6.3, 7.2, 8.5, 4.2, 6.3, 3.2, 5.0, 4.9, 6.6]
# Compute the mean and standard deviation
xbar = np.mean(lifetime)
s = np.std(lifetime)
## calculate 95% confidence z-interval
n = len(lifetime)
zstar95 = stats.norm.interval(0.95)
low = xbar + zstar95[0]*s/math.sqrt(n)
high = xbar + zstar95[1]*s/math.sqrt(n)
print("95% confidence z-interval: [",round(low,2),",", round(high,2),"] hours")
## calculate 90% confidence z-interval
n = len(lifetime)
zstar90 = stats.norm.interval(0.9)
low = xbar + zstar90[0]*s/math.sqrt(n)
high = xbar + zstar90[1]*s/math.sqrt(n)
print("90% confidence z-interval: [",round(low,2),",", round(high,2),"] hours")
## calculate 99% confidence z-interval
n = len(lifetime)
zstar99 = stats.norm.interval(0.99)
low = xbar + zstar99[0]*s/math.sqrt(n)
high = xbar + zstar99[1]*s/math.sqrt(n)
print("99% confidence z-interval: [",round(low,2),",", round(high,2),"] hours")
95% confidence z-interval: [ 5.0 , 7.13 ] hours
90% confidence z-interval: [ 5.17 , 6.96 ] hours
99% confidence z-interval: [ 4.66 , 7.46 ] hours
Home Activity
Recalculate the confidence intervals using the t-distribution.
# Add your solution here
13.4.6. Important Assumptions#
Student’s t-distribution only applies if the population is normally distributed and samples are random (i.e., zero covariance). Otherwise, the significance level is not correct, i.e., we will either under- or over-estimate uncertainty.
How to know/check if the population is normally distributed?
Preferred: Examine large amounts of historical data or leverage additional knowledge.
Practical: Plot the sample and check for outliers.
plt.hist(lifetime)
plt.title("Histogram of Catalyst Lifetime")
plt.xlabel("Lifetime [hours]")
plt.ylabel("Frequency")
plt.show()

Note
Class Discussion: Are there outliers in the catalyst lifetime dataset?
another_example = [1.2, 2.3, 1.5, 1.4, 1.3, 1.6, 1.4, 1.7, 1.6, 1.5, 1.6]
plt.hist(another_example,color="red")
plt.title("Another Example")
plt.xlabel("Measured Variable")
plt.ylabel("Frequency")
plt.show()
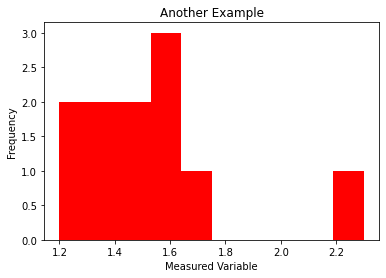
Note
Class Discussion: What about this dataset? Does it contain any outliers?
Another Example: Time Series Data.
Below are two plots of yield versus time data for two chemical processes.
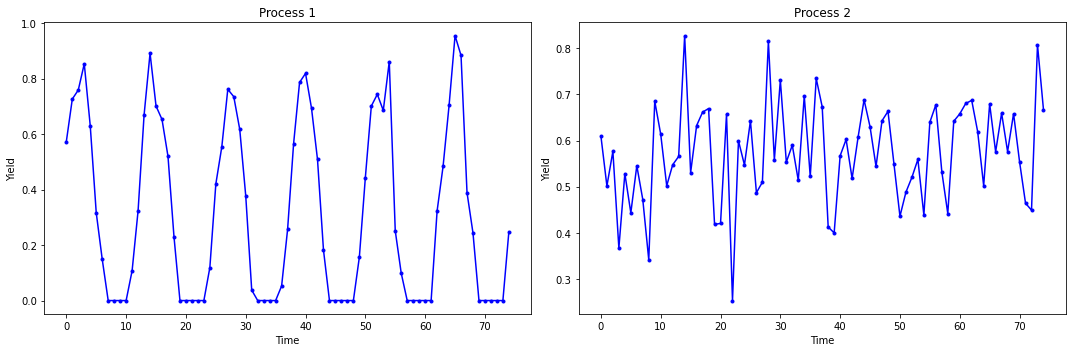
Note
Class Discussion: Can you use these data to calculate confidence intervals for the average yield of each process using a t-distribution?

