4.3. Invertible Matrix Theorem and Gaussian Elimination Example#
Reference: Chapter 7 of Computational Nuclear Engineering and Radiological Science Using Python, R. McClarren (2018)
4.3.1. Learning Objectives#
After studying this notebook, completing the activities, and asking questions in class, you should be able to:
Explain the Invertible Matrix Theorem to a classmate.
Solve mass balance problems with linear algebra and Gaussian elimination.
import numpy as np
import matplotlib.pyplot as plt
#this line is only needed in Jupyter notebooks
%matplotlib inline
4.3.2. Linear Algebra: Invertible Matrix Theorem#
Please review The Invertible Matrix Theorem in your linear algebra textbook (by Lay et al).
To paraphrase, \( A \) is a square \(n \times n\) matrix. The following statements are equivalent (all true or all false):
\(A\) is invertible
\(A\) is row equivalent to the \(n \times n\) identify matrix
\(A\) has \(n\) pivot positions
\(A x = 0\) has only the trivial solution
The columns of \(A\) form a linearly independent set
The system \(A x = b\) has at least one solution
Columns of \(A\) span \(\mathbb{R}^n\)
\(A^T\) is invertible
The rank of \(A\) is \(n\)
The determinant of \(A\) is not zero
4.3.3. Mass Balance Example (Adapted from an Old Exam Problem)#
Let’s see how Gaussian elimination can be applied to mass balance problems.
We want to convert A into valuable product B using a process with a mixer, reactor, and separator.
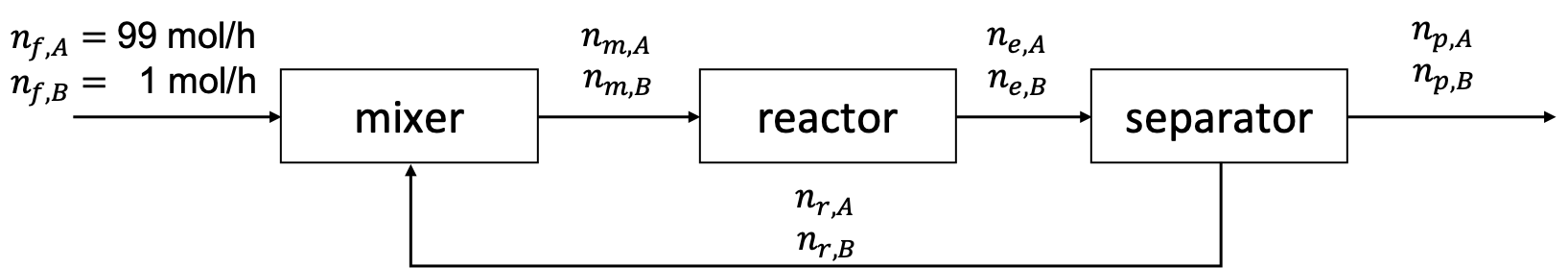
A feed enters at a rate of 100 moles per hour and is composed 99 mole % A and the remainder B. Unfortunately, the reactor only has a 10% per pass conversion efficiency. This means that only 10% of A that enters the reactor undergoes the reaction \(A \rightarrow B\). The separator splits the reactor effluent into two streams: the product and the recycle. The separator operates such that for every 1 mole of A in the product streams, there are 4 moles of A in the recycle stream. Likewise, for every 8 moles of B in the product stream, there is 1 mole of B in the recycle stream.
As shown in the figure above, we can model the process using component molar flowrates as variables. The first subscript is the stream (\(m\), \(e\), \(r\), or \(p\)) and the second subscript is the component (\(A\) or \(B\)).
4.3.4. Mathematical Model (Old Exam Question)#
Your classmate proposes the following mathematical model
Equation A: \(n_{f,A} + n_{r,A} = n_{m,A}\)
Equation B: \(n_{f,B} + n_{r,B} = n_{m,B}\)
Equation C: \(0.9 ~ n_{m,A} = n_{e,A}\)
Equation D: \(n_{m,A} + n_{m,B} = n_{e,A} + n_{e,B}\)
Equation E: \(n_{e,A} = n_{p,A} + n_{r,A}\)
Equation F: \(n_{e,B} = n_{p,B} + n_{r,B}\)
Equation G: \(4 ~ n_{p,A} = n_{r,A}\)
Equation H: \(n_{p,B} = 8 ~ n_{r,B}\)
Multiple choice description options:
Entire Process: Overall mole balance
Entire Process: Component A mole balance
Process: Component B mole balance
Mixer: Overall mole balance
Mixer: Component A mole balance
Mixer: Component B mole balance
Mixer: Summation equation
Reactor: Overall mole balance
Reactor: Component A mole balance
Reactor: Component B mole balance
Reactor: 90% of A fed into reactor is converted to B
Reactor: 10% of A fed into reactor is converted to B
Separator: Overall mole balance
Separator: Component A mole balance
Separator: Component B mole balance
Separator: For every 4 moles of A in recycle, there are 1 moles of A in product
Seperator: For every 1 mole of A in recycle, there are 4 moles of A in product
Seperator: For every 8 moles of B in recycle, there are 1 moles of B in product
Seperator: For every 1 mole of B in recycle, there are 1 moles of B in product
None of the above
Hint: Choice 19 does not include a typo. It is meant to be 1 and 1.
Home Activity
Match each equation to a description above. Record your answer in the dictionary eqn_names using the keys A,…,F and the values 1,…,20.
eqn_names = {'A':0, 'B':0, 'C':0, 'D':0, 'E':0, 'F':0, 'G':0, 'H':0}
# Add your solution here
# Removed autograder test. You may delete this cell.
4.3.5. Matrix Form (Old Exam Question)#
Class Activity
Convert the model to matrix form.
4.3.6. Solve with Gaussian Elimination#
def print_solution(x):
'''Print solution for this specific mole balance problem
Argument:
x: solution vector
Returns:
Nothing
'''
print("Stream description : Component = flowrate")
print("mixer outlet: A = ",x[0])
print("mixer effluent: B = ",x[1])
print("reactor effluent: A = ",x[2])
print("reactor effluent: B = ",x[3])
print("product: A = ",x[4])
print("product: B = ",x[5])
print("recycle: A = ",x[6])
print("recycle: B = ",x[7])
Class Activity
Finish populating A_ex and b_ex below.
## Assemble A matrix
A_ex = np.zeros((8,8))
A_ex[0,0] = 1
A_ex[0,6] = -1
A_ex[1,1] = 1
A_ex[1,7] = -1
# Add your solution here
A_ex[6,4] = 4
A_ex[6,6] = -1
A_ex[7,5] = 1
A_ex[7,7] = -8
print("A=\n",A_ex)
## Assemble b vector
b_ex = np.zeros(8)
b_ex[0] = 99
# Add your solution here
print("\nb=\n",b_ex)
Run the following cell with the Gaussian Elimination code developed in the last notebook.
def GaussElim(A,b,LOUD=False):
"""create a Gaussian elimination matrix for a system
Args:
A: N by N array
b: array of length N
Returns:
augmented matrix ready for back substitution
"""
# Extract dimensions of A
[Nrow, Ncol] = A.shape
# Check that A is square
assert Nrow == Ncol
# Check dimensions of b
N = Nrow
assert b.size == N
# create augmented matrix, just like
# code at the start of motivating example
aug_matrix = np.zeros((N,N+1))
aug_matrix[0:N,0:N] = A.copy()
aug_matrix[:,N] = b.copy()
#augmented matrix is created
if LOUD:
print("Augmented Matrix = ",aug_matrix)
# loop over columns
for column in range(0,N):
# loop over rows
for row in range(column+1,N):
# select row to modify
mod_row = aug_matrix[row,:]
# modify entire row
mod_row -= (mod_row[column]/aug_matrix[column,column]*
aug_matrix[column,:])
# overwrite row. Is this needed? Why or why not?
aug_matrix[row] = mod_row
# end loop over rows
# end loop over columns
# return augmented matrix
return aug_matrix
def swap_rows(A, a, b):
"""Rows two rows in a matrix, switch row a with row b
args:
A: matrix to perform row swaps on
a: row index of matrix
b: row index of matrix
returns: nothing
side effects:
changes A to rows a and b swapped
"""
# A negative index will give unexpected behavior
assert (a>=0) and (b>=0)
N = A.shape[0] #number of rows
assert (a<N) and (b<N) #less than because 0-based indexing
# create a temporary variable to store row a
temp = A[a,:].copy()
# move row b values into the location of row a
A[a,:] = A[b,:].copy()
# move row a values (stored in temp) into the location of row a
A[b,:] = temp.copy()
def BackSub(aug_matrix,z_tol=1E-8):
"""back substitute a N by N system after Gauss elimination
Args:
aug_matrix: augmented matrix with zeros below the diagonal [numpy 2D array]
z_tol: tolerance for checking for zeros below the diagonal [float]
Returns:
x: length N vector, solution to linear system [numpy 1D array]
"""
[Nrow, Ncol] = aug_matrix.shape
try:
# check the dimensions
assert Nrow + 1 == Ncol
except AssertionError:
print("Dimension checks failed.")
print("Nrow = ",Nrow)
print("Ncol = ",Ncol)
raise
assert type(z_tol) is float, "z_tol must be a float"
# check augmented matrix is all zeros below the diagonal
for r in range(Nrow):
for c in range(0,r):
assert np.abs(aug_matrix[r,c]) < z_tol, "\nWarning! Nonzero in position "+str(r)+","+str(c)
# create vector of zeros to store solution
x = np.zeros(Nrow)
# loop over the rows starting at the bottom
for row in range(Nrow-1,-1,-1):
RHS = aug_matrix[row,Nrow] # far column
# loop over the columns to the right of the diagonal
for column in range(row+1,Nrow):
# subtract, i.e., substitute the already known values
RHS -= x[column]*aug_matrix[row,column]
# compute the element of x corresponding to the current row
x[row] = RHS/aug_matrix[row,row]
return x
def GaussElimPivotSolve(A,b,LOUD=False):
"""create a Gaussian elimination with pivoting matrix for a system
Args:
A: N by N array
b: array of length N
Returns:
solution vector in the original order
"""
# checks dimensions
[Nrow, Ncol] = A.shape
assert Nrow == Ncol
N = Nrow
#create augmented matrix
aug_matrix = np.zeros((N,N+1))
aug_matrix[0:N,0:N] = A
aug_matrix[:,N] = b
#augmented matrix is created
#create scale factors
s = np.zeros(N)
count = 0
for row in aug_matrix[:,0:N]: #don't include b
s[count] = np.max(np.fabs(row))
count += 1
# print diagnostics if requested
if LOUD:
print("s =",s)
print("Original Augmented Matrix is\n",aug_matrix)
#perform elimination
for column in range(0,N):
## swap rows if needed
# find largest position
largest_pos = (np.argmax(np.fabs(aug_matrix[column:N,column]/
s[column:N])) + column)
# check if current column is largest position
if (largest_pos != column):
# if not, swap rows
if (LOUD):
print("Swapping row",column,"with row",largest_pos)
print("Pre swap\n",aug_matrix)
swap_rows(aug_matrix,column,largest_pos)
#re-order s
tmp = s[column]
s[column] = s[largest_pos]
s[largest_pos] = tmp
if (LOUD):
print("A =\n",aug_matrix)
print("new_s =\n", s)
#finish off the row
for row in range(column+1,N):
mod_row = aug_matrix[row,:]
mod_row = mod_row - mod_row[column]/aug_matrix[column,column]*aug_matrix[column,:]
aug_matrix[row] = mod_row
#now back solve
if LOUD:
print("Final aug_matrix is\n",aug_matrix)
x = BackSub(aug_matrix)
return x
## Solve
x = GaussElimPivotSolve(A_ex, b_ex, LOUD=True)
print_solution(x)
4.3.7. Flip the order of the row#
What happens if we write the equations in reserve order?
A_flip = np.flipud(A_ex.copy())
b_flip = np.flipud(b_ex.copy())
print("A=\n",A_flip)
print("\nb=\n",b_flip)
A=
[[ 0. 0. 0. 0. 0. 1. 0. -8. ]
[ 0. 0. 0. 0. 4. 0. -1. 0. ]
[ 0. 0. 0. 1. 0. -1. 0. -1. ]
[ 0. 0. 1. 0. -1. 0. -1. 0. ]
[ 1. 1. -1. -1. 0. 0. 0. 0. ]
[ 0.9 0. -1. 0. 0. 0. 0. 0. ]
[ 0. 1. 0. 0. 0. 0. 0. -1. ]
[ 1. 0. 0. 0. 0. 0. -1. 0. ]]
b=
[ 0. 0. 0. 0. 0. 0. 1. 99.]
x_flip = GaussElimPivotSolve(A_flip, b_flip, LOUD=True)
print_solution(x_flip)
We see that we get the same order but need to pivot! Let’s try without pivoting.
aug_mat = GaussElim(A_flip,b_flip)
print("\naug_mat = \n",aug_mat)
aug_mat =
[[ 0. 0. 0. 0. 0. 1. 0. -8. 0.]
[nan nan nan nan nan nan nan nan nan]
[nan nan nan nan nan nan nan nan nan]
[nan nan nan nan nan nan nan nan nan]
[nan nan nan nan nan nan nan nan nan]
[nan nan nan nan nan nan nan nan nan]
[nan nan nan nan nan nan nan nan nan]
[nan nan nan nan nan nan nan nan nan]]
/var/folders/3w/vr4xmyqs451dg23xk88pqcg00000gq/T/ipykernel_47904/826583952.py:40: RuntimeWarning: invalid value encountered in double_scalars
mod_row -= (mod_row[column]/aug_matrix[column,column]*
/var/folders/3w/vr4xmyqs451dg23xk88pqcg00000gq/T/ipykernel_47904/826583952.py:40: RuntimeWarning: divide by zero encountered in double_scalars
mod_row -= (mod_row[column]/aug_matrix[column,column]*
/var/folders/3w/vr4xmyqs451dg23xk88pqcg00000gq/T/ipykernel_47904/826583952.py:40: RuntimeWarning: invalid value encountered in multiply
mod_row -= (mod_row[column]/aug_matrix[column,column]*
For this problem, we must use pivoting if we start with the equations in a certain order.
4.3.8. Replace the 8th equation with an overall mole balance#
A_new = A_ex.copy()
b_new = b_ex.copy()
A_new[7,:] = np.zeros(8)
A_new[7,4] = 1
A_new[7,5] = 1
b_new[7] = 100
print("A=\n",A_new)
print("\nb=\n",b_new)
A=
[[ 1. 0. 0. 0. 0. 0. -1. 0. ]
[ 0. 1. 0. 0. 0. 0. 0. -1. ]
[ 0.9 0. -1. 0. 0. 0. 0. 0. ]
[ 1. 1. -1. -1. 0. 0. 0. 0. ]
[ 0. 0. 1. 0. -1. 0. -1. 0. ]
[ 0. 0. 0. 1. 0. -1. 0. -1. ]
[ 0. 0. 0. 0. 4. 0. -1. 0. ]
[ 0. 0. 0. 0. 1. 1. 0. 0. ]]
b=
[ 99. 1. 0. 0. 0. 0. 0. 100.]
x_new = GaussElimPivotSolve(A_new, b_new, LOUD=True)
Why did this fail? Especially because, 1 mole of A converts to 1 mole of B, we know the overall mole balance is a valid equation.
Class Activity
Discuss with a partner. Hint: How is the invertible matrix theorem helpful?
# hint: find the rank of A_ex
# Add your solution here
# hint: find the rank of A_new
# Add your solution here

