Multi-Objective Optimization#
Prepared by: Shammah Lilonfe (slilonfe@nd.edu, 2024) and Zhicheng Lu (zlu3@nd.edu, 2024) at the University of Notre Dame.
Notes for edits:
Add standard imports so this runs on Colab and locally
Remove material in prior notebook
Verify notebook runs
Adjust equation numbering
import sys
if "google.colab" in sys.modules:
!wget "https://raw.githubusercontent.com/ndcbe/optimization/main/notebooks/helper.py"
import helper
helper.install_idaes()
helper.install_ipopt()
else:
sys.path.insert(0, '../')
import helper
# Loads IPOPT and other solvers
import idaes
helper.set_plotting_style()
# Importing packages to be used in this project
import pyomo.environ as pyo
import pyomo.dae as dae
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
Learning Objectives#
Explain the modelling strategies of multi-objective optimization (MOO)
Demonstrate the power of MOO in solving chemical industry problems
Basic Definitions#
Let \(\mathbf{x} \in \mathbb{R}^{n_x}\) be the decision vector that lies in the feasible set \(X \subseteq \mathbb{R}^{n_x}\), which is assumed to be compact and nonempty. Given \(n\) objective functions \(f_i: X \to \mathbb{R}\) for \(i \in O:\) = {1, 2, \(\dots, n\)}, and the corresponding objective vector: \(\mathbf{f}(\mathbf{x}) = \big(f_1(\mathbf{x}), f_2(\mathbf{x}), \dots, f_{n}(\mathbf{x})\big)\). Assuming that the objective functions remain bounded in \(X\).
The MOO problem is defined as: $\(\begin{equation} \min_{x \in X} \big(f_1(x), f_2(x), \dots, f_{n}(x)\big) \tag{1.1} \end{equation}\)$
Again, we assume that the minimization operand implies global minimization and that the objective functions have been scaled so that their values lie in the interval [0, 1].
This was taken from Dowling et al. (2016).
Scaling#
Scaling of the objective functions is performed using the coordinates of the utopia and nadirpoints as follows. We define:
Here, the coordinates of the utopia point are given by \(\underline{f}_i\), while those of the nadir point are given by:
The objectives are scaled as follows:
We can prevent visiting regions beyond the nadir points by imposing the constraints:
This was taken from Dowling et al. (2016).
Pareto Optimality#
Below are the standard definitions of weak Pareto optimality and Pareto optimality (Miettinen, 1999):
Definition 1 (Weak Pareto Optimality): A decision \(x^*\) with objectives \(f_i(x^*), \, i \in O\) is a weak Pareto solution of MOO if there does not exist an alternate solution \(x\) with objectives \(f_i(x), \, i \in O\) which satisfies \(f_i(x) < f_i(x^*)\) for all \(i \in O\).
Definition 2 (Pareto Optimality): A decision \(x^*\) with objectives \(f_i(x^*), \, i \in O\) is a Pareto solution of MOO if there does not exist an alternate solution \(x\) with objectives \(f_i(x), \, i \in O\) which satisfies \(f_i(x) \leq f_i(x^*)\) for all \(i \in O\) and at least one index \(i\) satisfies \(f_i(x) < f_i(x^*)\).
Any Pareto solution of MOO is a weak Pareto solution. To see this, we consider the following contradiction: assume \(x^*\) is a Pareto solution, and thus no alternate decision \(x\) exists such that \(f_i(x) \leq f_i(x^*)\) for all \(i \in O\) and \(f_i(x) < f_i(x^*)\) for at least one index \(i\). If, in addition, we assume that \(x^*\) is not weak Pareto, then there exists an alternate decision \(x\) such that \(f_i(x) < f_i(x^*)\) for all \(i \in O\). This is a contradiction, and thus \(x^*\) must be a Pareto and a weak Pareto solution.
The set of all Pareto solutions for MOO is known as the Pareto set. There are different methods that can be used to compute elements of the Pareto set, such as the weighting method. The results associated with this method will become relevant in the following sections.
Consider a weight vector \(\mathbf{w} \in \mathbb{R}^n\) and assume that this satisfy the conditions:
In some cases, we will require the weights to satisfy the stronger condition:
Consider now the weighted problem:
The solutions of this problem satisfy the following property:
Proposition 1: Let \(x^*\) be a solution of the weighted problem (1.8):
If the weight vector \(\mathbf{w}\) satisfies \(w_i \geq 0, \, i \in O\) (Condition 1.6a), then \(x^*\) is a weak Pareto solution of MOO.
If the weight vector \(\mathbf{w}\) satisfies \(w_i > 0, \, i \in O\) (Condition 1.7a), then \(x^*\) is a Pareto solution of MOO.
The nadir point is obtained by computing worst-case value results of the utopia point as in (1.2) and (1.3). The nadir point coordinates are determined by minimizing each objective function and then obtaining the corresponding worst-case values for the objectives. This approach is equivalent to solving (1.1) with \(w_i=1\) and \(w_{i' \neq i}=0\) for \(i, i' ∈ O\). From Proposition 1, these single objective solutionsare only weak Pareto solutions. Consequently, it is possible to achieve a further reduction in the other objectives without compromising the original single objective. Because of this, the nadir point may unnecessarily enlarges the decision space. This inturn impacts the scaling of the objectives in (1.4). To address this issue, an alternate nadir point is exploited.
Definition 3 (Alternate Nadir Point): Let \(\underline{x}_i\) and \(\underline{f}_i\) be defined as in (1.2) for all \(i \in O\). For each \(i \in O\), let the alternate nadir solution \(\underline{x}^*_i\) be defined as:
where \(\mathbf{w}\) satisfies \(w_i = 0\) and \(w_{i' \neq i} > 0\).
The alternate nadir point \(\mathbf{\bar{f}}^*\) with elements \(\bar{f}^*_i\) is defined as:
Proposition 2: The alternate nadir solutions \(\underline{x}^*_i\) for \(i \in O\), are Pareto solutions of MOO.
The alternate nadir point method requires a selection of weights for the objective functions. Such a selection is not unique and thus different alter-nate nadir solutions can be obtained. However, all of these alternatives are guaranteed to be Pareto solutions. Even small nonzero weights guarantee this property and thus the alternate nadir point can be designed to prevent extreme degeneracies of the objectives resulting from the standard use of (1.2) and (1.3).
Computing all the elements in the the Pareto set is computationally intractable, particularly when many objectives are considered. It is well known, however, that one can compute Pareto optimal compromise solutions by finding points that are closest in distance to the utopia point. This is often done by solving the minimization problem:
where \(\|\cdot\|_p\) is the \(L_p\) norm.
Proposition 3. A solution \(x^*\) of the minimum distance problem (1.11) is a Pareto solution of MOO.
This was taken from Dowling et al. (2016).
Epsilon-Constraint Method#
Key Concepts#
In MOO, the goal is to minimize a vector of objective functions:
Subject to: $\(\begin{equation} g_j(x) \leq 0, \quad j = 1, \dots, m. \tag{2.1} \end{equation}\)$
Where \(f_1, f_2, \dots, f_n\) are the objectives, and \(g_j(x)\) are problem-specific constraints.
The epsilon-constrained method reformulates this problem by optimizing one objective function (e.g., \(f_1(x)\)), while treating the remaining objectives as constraints with thresholds \(\epsilon_i\) for \(i \in O:\) = {1, 2, \(\dots, n\)}. The reformulated problem is expressed as:
Subject to: $\(\begin{equation} f_2(x) \leq \epsilon_2, \quad f_3(x) \leq \epsilon_3, \quad \dots, \quad f_n(x) \leq \epsilon_n, \end{equation}\)\( \)\(\begin{equation} g_j(x) \leq 0, \quad j = 1, \dots, m. \tag{2.2} \end{equation}\)$
Adopted from Miettinen (1999).
Key Points#
Trade-Off Exploration: By systematically varying the values of \(\epsilon_2, \epsilon_3, \dots, \epsilon_n\), it is possible to explore trade-offs between objectives and construct an approximation of the Pareto front.
Reduction to Single Objective: The epsilon-constrained method simplifies the multi-objective problem into a single-objective problem for each choice of \(\epsilon\) values.
Flexibility: Any one of the objectives can be chosen as the primary objective to optimize, allowing the user to prioritize specific goals.
Miettinen (1999).
Example: Design of Chemical Reactors for Chemical Transformations#
Chemical reactors are often the most important unit operations in a chemical plant. Reactors come in many forms, however two of the most common idealizations are the continuously stirred tank reactor (CSTR) and the plug flow reactor. The CSTR is often used in modeling studies, and it can be effectively modeled as a lumped parameter system. In this example, we will consider the following reaction scheme known as the Van de Vusse reaction:
A diagram of the system is shown in figure below, where F is the volumetric flowrate. The reactor is assumed to be filled to a constant volume, and the mixture is assumed to have constant density, so the volumetric flowrate into the reactor is equal to the volumetric flowrate out of the reactor. Since the reactor is assumed to be wellmixed, the concentrations in the reactor are equivalent to the concentrations of each component flowing out of the reactor, given by \(C_A\), \(C_B\), \(C_C\), and \(C_D\).
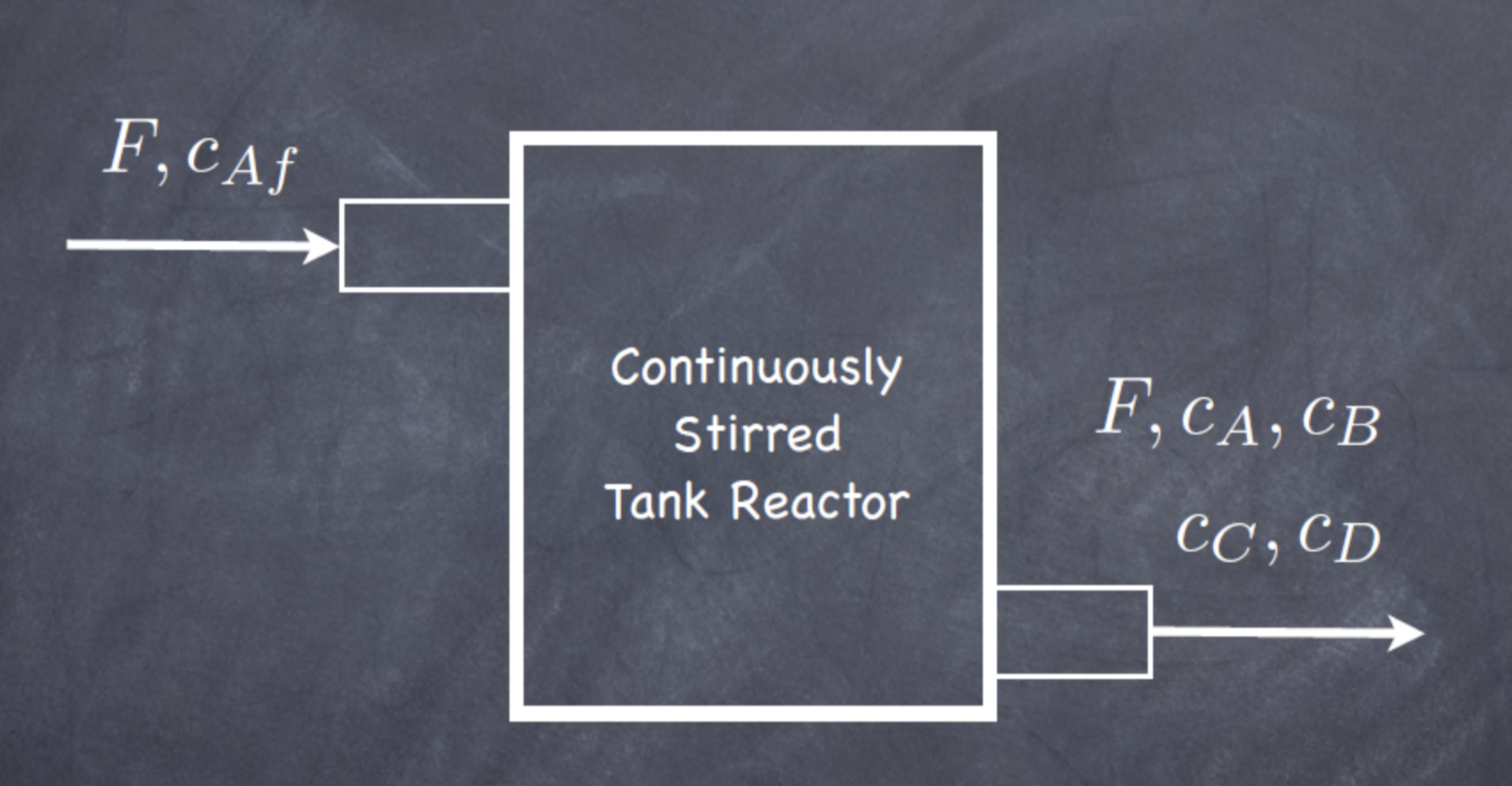
This example (both text and figure) was taken from Hart et al (2017).
See Chemical Reactor Design for the extension of this example to design a batch reactor. This notebook extends the batch reactor problem into a multi-objective optimization problem.
Multi-Objective Optimization to Maximize Species B and C#
In this project, species B and C are the desired products, and we are considering the dynamic state of the system (most often, industrial operations are dynamic). The goal of this work is to determine the volume of the reactor that will maximize the concentration of species B and C in the effluent after 10 minutes. Furthermore, for economic reasons, the reactor volume should not exceed \(\mathrm{100\,m^3}\).
The mole balance for each of the species in this system under dynamic state is given by:
Where \(C_A\), \(C_B\), \(C_C\) and \(C_D\) are the concentrations of species A, B, C and D, respectively. \(F\) is the feed flow rate, \(C_{Af}\) is the concentration of species A in the feed, \(k_1\), \(k_2\) and \(k_3\) are the rate constants of reaction 1 (species A to B), reaction 2 (species B to C) and reaction 3 (species A to D), respectively.
The data for the system are: $\(\begin{align*} C_{Af}=10^4\,\mathrm{\frac{mol}{m^3}},\quad{k_1=\frac{5}{6}\,\mathrm{min^{-1}}},\quad{k_2=\frac{5}{3}\,\mathrm{min^{-1}}},\quad{k_3=\frac{1}{6000}\,\mathrm{\frac{m^3}{mol\cdot min}}},\quad{F=1\,\mathrm{\frac{m^3}{min}}} \end{align*}\)$
The data was obtained from Hart et al (2017).
Optimization Model Formulation#
Mathematically, we want to solve the following optimal reactor volume problem:
Degrees of Freedom Analysis#
From the optimization model formulated above:
Number of variables = 5
Number of equality constraints = 4
Therefore, the degrees of freedom = 5 - 4 = 1
Weighted Method Pyomo Implementation#
# parameters
conc_A_feed = 10000 # mol/m^3
k1 = 5 / 6 # min^-1
k2 = 5 / 3 # min^-1
k3 = 1 / 6000 # m^3/mol/min
feed_flowrate = 1 # m^3/min
# Function for weighted method multi-objective dynamic model
def weighted_reactor_model(F, CAF, k1, k2, k3, w1, w2):
"""Creates reactor model with multi-objective optimization
Arguments:
F: feed volumetric flow rate in m^3/min
CAF: feed concentration of A, gmol/m^3
k1: rate constant for A to B, min^-1
k2: rate constant for B to C, min^-1
k3: rate constant for 2A to D, m^3/(gmol.min)
w1: weights for objective 1
w2: weights for objective 2
Returns:
model: pyomo model
"""
# Create Pyomo model
model = pyo.ConcreteModel()
# Define time as a continuous set
model.t = dae.ContinuousSet(bounds=(0, 10)) # Time from 0 to 10 minutes
# Define variables
model.CA = pyo.Var(model.t, within=pyo.NonNegativeReals) # Concentration of A
model.CB = pyo.Var(model.t, within=pyo.NonNegativeReals) # Concentration of B
model.CC = pyo.Var(model.t, within=pyo.NonNegativeReals) # Concentration of C
model.CD = pyo.Var(model.t, within=pyo.NonNegativeReals) # Concentration of D
model.V = pyo.Var(within=pyo.NonNegativeReals, bounds=(0, 100), initialize=11.35) # Reactor volume
# Define derivatives
model.dCA_dt = dae.DerivativeVar(model.CA, wrt=model.t)
model.dCB_dt = dae.DerivativeVar(model.CB, wrt=model.t)
model.dCC_dt = dae.DerivativeVar(model.CC, wrt=model.t)
model.dCD_dt = dae.DerivativeVar(model.CD, wrt=model.t)
# CA differential equation
def ca_rate_ode(m, t):
return m.dCA_dt[t] == (F / m.V) * CAF - (F / m.V) * m.CA[t] - k1 * m.CA[t] - 2 * k3 * m.CA[t] ** 2
model.CA_ode = pyo.Constraint(model.t, rule=ca_rate_ode)
# CB differential equation
def cb_rate_ode(m, t):
return m.dCB_dt[t] == -(F / m.V) * m.CB[t] + k1 * m.CA[t] - k2 * m.CB[t]
model.CB_ode = pyo.Constraint(model.t, rule=cb_rate_ode)
# CC differential equation
def cc_rate_ode(m, t):
return m.dCC_dt[t] == -(F / m.V) * m.CC[t] + k2 * m.CB[t]
model.CC_ode = pyo.Constraint(model.t, rule=cc_rate_ode)
# CD differential equation
def cd_rate_ode(m, t):
return m.dCD_dt[t] == -(F / m.V) * m.CD[t] + k3 * m.CA[t] ** 2
model.CD_ode = pyo.Constraint(model.t, rule=cd_rate_ode)
# Initial conditions
def init_conditions(m):
yield m.CA[0] == CAF
yield m.CB[0] == 0
yield m.CC[0] == 0
yield m.CD[0] == 0
model.init_conditions = pyo.ConstraintList(rule=init_conditions)
# Multi-objective function: weighted sum of objectives
# Maximize CC (product C), CB (intermediate B)
def objective_function(m):
return w1 * m.CC[10] + w2 * m.CB[10]
model.objective = pyo.Objective(rule=objective_function, sense=pyo.maximize)
return model
# Weights for multi-objective function
weights = {"w1": 0.5, "w2": 0.5}
# Create reactor model
reactor_model = weighted_reactor_model(feed_flowrate, conc_A_feed, k1, k2, k3, weights["w1"], weights["w2"])
# Discretize the model using collocation
discretizer = pyo.TransformationFactory('dae.collocation')
discretizer.apply_to(reactor_model, nfe=20, ncp=10)
# Solve the model
solver = pyo.SolverFactory('ipopt')
results = solver.solve(reactor_model, tee=True)
# Print results
optimal_volume = pyo.value(reactor_model.V)
optimal_CC = pyo.value(reactor_model.CC[10]) * 1e-3 # kmol/m^3
optimal_CB = pyo.value(reactor_model.CB[10]) * 1e-3 # kmol/m^3
print(f"Optimal reactor volume: {round(optimal_volume, 2)} m^3")
print(f"Maximum Concentration of C at optimal reactor volume is: {round(optimal_CC, 2)} kmol/m^3")
print(f"Maximum Concentration of B at optimal reactor volume is: {round(optimal_CB, 2)} kmol/m^3")
# Extract results for plotting
time = [t for t in reactor_model.t] # minutes
CA_values = [(pyo.value(reactor_model.CA[t])*10**(-3)) for t in time] # kmol/m^3
CB_values = [(pyo.value(reactor_model.CB[t])*10**(-3)) for t in time] # kmol/m^3
CC_values = [(pyo.value(reactor_model.CC[t])*10**(-3)) for t in time] # kmol/m^3
CD_values = [(pyo.value(reactor_model.CD[t])*10**(-3)) for t in time] # kmol/m^3
# Plot the results
plt.figure(figsize=(8, 6))
plt.plot(time, CA_values, label='$\mathrm{C_{A}}$')
plt.plot(time, CB_values, label='$\mathrm{C_{B}}$')
plt.plot(time, CC_values, label='$\mathrm{C_{C}}$')
plt.plot(time, CD_values, label='$\mathrm{C_{D}}$')
plt.xlabel('Time (min)', fontsize=16,fontweight='bold')
plt.ylabel('Concentration (kmol/$\mathbf{m^3}$)', fontsize=16,fontweight='bold')
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.title('Concentration Profiles Over Time', fontsize=16,fontweight='bold')
plt.legend()
plt.show()
Ipopt 3.13.2:
******************************************************************************
This program contains Ipopt, a library for large-scale nonlinear optimization.
Ipopt is released as open source code under the Eclipse Public License (EPL).
For more information visit http://projects.coin-or.org/Ipopt
This version of Ipopt was compiled from source code available at
https://github.com/IDAES/Ipopt as part of the Institute for the Design of
Advanced Energy Systems Process Systems Engineering Framework (IDAES PSE
Framework) Copyright (c) 2018-2019. See https://github.com/IDAES/idaes-pse.
This version of Ipopt was compiled using HSL, a collection of Fortran codes
for large-scale scientific computation. All technical papers, sales and
publicity material resulting from use of the HSL codes within IPOPT must
contain the following acknowledgement:
HSL, a collection of Fortran codes for large-scale scientific
computation. See http://www.hsl.rl.ac.uk.
******************************************************************************
This is Ipopt version 3.13.2, running with linear solver ma27.
Number of nonzeros in equality constraint Jacobian...: 12619
Number of nonzeros in inequality constraint Jacobian.: 0
Number of nonzeros in Lagrangian Hessian.............: 1006
Total number of variables............................: 1609
variables with only lower bounds: 804
variables with lower and upper bounds: 1
variables with only upper bounds: 0
Total number of equality constraints.................: 1608
Total number of inequality constraints...............: 0
inequality constraints with only lower bounds: 0
inequality constraints with lower and upper bounds: 0
inequality constraints with only upper bounds: 0
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
0 -9.9999900e-03 1.00e+04 1.93e+00 -1.0 0.00e+00 - 0.00e+00 0.00e+00 0
1 -6.9106248e-03 1.00e+04 1.72e+00 -1.0 1.06e+04 - 9.90e-07 1.71e-06f 2
2 -5.1191593e-03 1.00e+04 1.95e+00 -1.0 1.03e+04 - 2.69e-06 1.96e-06f 2
3 -4.7030315e-03 1.00e+04 2.43e+00 -1.0 1.01e+04 - 4.71e-06 1.37e-06f 3
4 -5.5812242e-03 1.00e+04 2.87e+00 -1.0 1.00e+04 - 2.41e-06 2.79e-06f 2
5 -2.7817761e-02 1.00e+04 4.64e+00 -1.0 1.00e+04 - 1.43e-06 1.31e-05f 1
6 -3.6134940e-02 1.00e+04 7.04e+00 -1.0 1.00e+04 - 1.22e-05 4.04e-06f 1
7 -3.9083441e-02 1.00e+04 2.17e+02 -1.0 1.00e+04 - 1.66e-05 1.43e-06f 1
8 -2.0478875e+03 3.18e+04 1.32e+07 -1.0 1.00e+04 - 1.93e-05 9.90e-01f 1
9 -2.0482528e+03 3.18e+04 1.32e+07 -1.0 3.25e+04 - 5.60e-05 4.99e-04f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
10 -2.0488906e+03 3.17e+04 1.32e+07 -1.0 3.25e+04 - 5.56e-04 8.72e-04f 1
11 -2.7715600e+03 3.11e+03 2.76e+06 -1.0 3.25e+04 - 1.45e-03 9.90e-01f 1
12 -2.7705317e+03 3.11e+03 2.75e+06 -1.0 1.81e+03 - 4.95e-01 1.85e-03h 1
13 -2.4623362e+03 1.38e+03 1.77e+06 -1.0 1.81e+03 - 5.56e-01 5.56e-01h 1
14 -2.3037964e+03 7.95e+02 1.25e+06 -1.0 8.25e+02 - 8.89e-01 4.16e-01h 1
15 -2.3030278e+03 7.93e+02 1.25e+06 -1.0 5.34e+02 - 8.54e-01 2.93e-03h 1
16 -2.3030202e+03 7.93e+02 3.61e+06 -1.0 5.33e+02 - 1.31e-01 2.93e-05h 1
17 -2.3030201e+03 7.93e+02 2.46e+06 -1.0 5.14e+02 - 3.40e-09 1.70e-07f 2
18 -2.3029949e+03 7.92e+02 1.85e+06 -1.0 4.66e+02 - 2.38e-08 2.40e-04f 1
19 -2.1978050e+03 1.29e+01 2.49e+06 -1.0 4.66e+02 - 2.37e-04 1.00e+00h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
20 -2.1854004e+03 8.65e-03 9.47e+04 -1.0 2.24e+01 - 9.90e-01 1.00e+00h 1
21 -2.1853877e+03 2.05e-08 9.54e+02 -1.0 2.19e-02 - 9.90e-01 1.00e+00f 1
22 -2.1853883e+03 1.67e-09 1.64e+01 -1.0 1.07e-03 - 9.83e-01 1.00e+00f 1
23 -2.1853906e+03 2.02e-08 3.84e+06 -2.5 3.74e-03 - 6.04e-01 1.00e+00f 1
24 -2.1867164e+03 7.06e-03 3.83e+06 -2.5 2.20e+00 - 4.26e-03 1.00e+00f 1
25 -2.2344141e+03 1.31e+01 7.12e+05 -2.5 7.90e+01 -4.0 8.14e-01 1.00e+00f 1
26 -2.4189753e+03 4.72e+02 4.52e+05 -2.5 3.02e+02 - 3.66e-01 1.00e+00f 1
27 -2.3520266e+03 2.24e+02 3.90e+01 -2.5 1.05e+02 -4.5 1.00e+00 1.00e+00h 1
28 -2.5066649e+03 6.35e+00 9.42e+00 -2.5 2.35e+02 - 1.00e+00 1.00e+00h 1
29 -2.7982612e+03 5.21e+04 3.60e+05 -2.5 4.54e+02 -5.0 1.00e+00 1.00e+00f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
30 -2.5091187e+03 4.80e+04 3.33e+05 -2.5 1.93e+04 -5.4 5.01e-02 6.96e-02h 4
31 -2.4432716e+03 4.77e+04 3.31e+05 -2.5 4.84e+04 -5.9 6.67e-02 6.59e-03h 6
32 -2.4271298e+03 4.77e+04 3.31e+05 -2.5 3.31e+05 - 1.08e-02 2.43e-04h 8
33 -2.4191049e+03 4.77e+04 3.31e+05 -2.5 2.59e+05 - 2.42e-02 1.55e-04h 9
34 -2.4181047e+03 4.77e+04 3.31e+05 -2.5 2.34e+05 - 3.22e-02 2.15e-05h 12
35 -1.9061686e+03 4.73e+04 3.24e+05 -2.5 2.31e+05 - 6.00e-02 1.11e-02h 3
36 -2.6199327e+02 5.58e+04 1.06e+05 -2.5 4.45e+04 - 1.00e+00 2.36e-01h 1
37 -4.1639345e+02 3.57e+04 6.20e+04 -2.5 1.10e+04 -5.5 6.54e-01 1.00e+00h 1
38 -5.2692143e+02 1.91e+04 3.40e+04 -2.5 3.94e+03 -5.1 4.11e-01 1.00e+00h 1
39 -1.0667580e+03 2.41e+04 2.12e+04 -2.5 1.23e+04 -5.5 3.54e-01 1.00e+00h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
40 -1.4537504e+03 1.35e+04 1.26e+04 -2.5 5.72e+03 -5.1 5.30e-01 1.00e+00h 1
41 -1.6697551e+03 1.26e+04 1.04e+04 -2.5 9.01e+03 -5.6 3.91e-01 2.18e-01h 1
42 -1.6690656e+03 1.26e+04 1.04e+04 -2.5 8.60e+03 - 7.03e-01 5.92e-04h 1
43 -1.6690328e+03 1.26e+04 1.04e+04 -2.5 3.60e+03 - 1.23e-03 9.46e-04h 1
44 -1.6685485e+03 1.24e+04 1.03e+04 -2.5 3.61e+03 - 1.15e-02 1.47e-02h 1
45 -1.6685458e+03 1.24e+04 1.03e+04 -2.5 3.57e+03 - 2.67e-04 7.86e-05h 1
46 -1.6681612e+03 1.22e+04 1.01e+04 -2.5 3.58e+03 - 9.87e-03 1.24e-02h 1
47 -1.6681596e+03 1.22e+04 1.01e+04 -2.5 3.54e+03 - 1.46e-04 5.05e-05h 1
48 -1.6679040e+03 1.21e+04 1.00e+04 -2.5 3.55e+03 - 7.74e-03 8.66e-03h 1
49 -1.6679022e+03 1.21e+04 1.00e+04 -2.5 3.53e+03 - 6.71e-05 6.23e-05h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
50 -1.6677659e+03 1.21e+04 1.00e+04 -2.5 3.53e+03 - 3.73e-03 4.77e-03h 1
51 -1.6677059e+03 1.20e+04 9.98e+03 -2.5 3.52e+03 - 1.19e-03 2.12e-03h 1
52 -1.6676318e+03 1.20e+04 9.95e+03 -2.5 3.52e+03 - 1.68e-03 2.62e-03h 1
53 -1.6676301e+03 1.20e+04 9.95e+03 -2.5 3.51e+03 - 2.83e-05 6.55e-05h 1
54 -1.6674758e+03 1.19e+04 9.90e+03 -2.5 3.51e+03 - 6.49e-04 5.72e-03h 1
55 -1.6674752e+03 1.19e+04 9.90e+03 -2.5 3.50e+03 - 7.19e-05 2.36e-05h 1
56 -1.6672799e+03 1.18e+04 9.82e+03 -2.5 3.50e+03 - 2.40e-03 7.80e-03h 1
57 -1.6672792e+03 1.18e+04 9.82e+03 -2.5 3.48e+03 - 1.19e-04 2.60e-05h 1
58 -1.6670827e+03 1.17e+04 9.74e+03 -2.5 3.49e+03 - 3.67e-04 8.64e-03h 1
59 -1.6670820e+03 1.17e+04 9.73e+03 -2.5 3.46e+03 - 2.17e-04 2.67e-05h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
60 -1.6669107e+03 1.16e+04 9.65e+03 -2.5 3.47e+03 - 9.95e-05 8.41e-03h 1
61 -1.6669102e+03 1.16e+04 9.65e+03 -2.5 3.45e+03 - 2.94e-04 2.54e-05h 1
62 -1.6667758e+03 1.16e+04 9.58e+03 -2.5 3.46e+03 - 1.02e-04 7.41e-03h 1
63 -1.6667754e+03 1.16e+04 9.58e+03 -2.5 3.43e+03 - 3.11e-04 2.23e-05h 1
64 -1.6666791e+03 1.15e+04 9.53e+03 -2.5 3.44e+03 - 2.96e-04 5.94e-03h 1
65 -1.6666788e+03 1.15e+04 9.53e+03 -2.5 3.42e+03 - 2.60e-04 1.82e-05h 1
66 -1.6666165e+03 1.14e+04 9.49e+03 -2.5 3.43e+03 - 8.23e-03 4.22e-03h 1
67 -1.6665798e+03 1.14e+04 9.46e+03 -2.5 3.42e+03 - 7.59e-04 2.66e-03h 1
68 -1.6665679e+03 1.14e+04 9.45e+03 -2.5 3.41e+03 - 4.86e-04 8.79e-04h 1
69 -1.6665512e+03 1.14e+04 9.44e+03 -2.5 3.41e+03 - 7.89e-04 1.25e-03h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
70 -1.6665138e+03 1.14e+04 9.41e+03 -2.5 3.41e+03 - 3.21e-04 2.98e-03h 1
71 -1.6665133e+03 1.14e+04 9.41e+03 -2.5 3.40e+03 - 6.82e-05 3.99e-05h 1
72 -1.6664672e+03 1.13e+04 9.37e+03 -2.5 3.40e+03 - 1.28e-03 4.05e-03h 1
73 -1.6664669e+03 1.13e+04 9.37e+03 -2.5 3.39e+03 - 6.39e-05 2.77e-05h 1
74 -1.6664203e+03 1.13e+04 9.33e+03 -2.5 3.39e+03 - 1.98e-03 4.66e-03h 1
75 -1.6664198e+03 1.13e+04 9.33e+03 -2.5 3.38e+03 - 5.84e-05 5.07e-05h 1
76 -1.6663796e+03 1.12e+04 9.29e+03 -2.5 3.38e+03 - 2.08e-03 4.70e-03h 1
77 -1.6663468e+03 1.11e+04 9.24e+03 -2.5 3.37e+03 - 6.84e-04 4.56e-03h 1
78 -1.6663247e+03 1.11e+04 9.21e+03 -2.5 3.36e+03 - 5.16e-04 3.65e-03h 1
79 -1.6663106e+03 1.11e+04 9.19e+03 -2.5 3.35e+03 - 3.55e-04 2.67e-03h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
80 -1.6663029e+03 1.11e+04 9.17e+03 -2.5 3.35e+03 - 2.44e-04 1.63e-03h 1
81 -1.6663016e+03 1.11e+04 9.17e+03 -2.5 3.34e+03 - 5.95e-05 2.82e-04h 1
82 -1.6662983e+03 1.10e+04 9.16e+03 -2.5 3.34e+03 - 2.63e-04 7.41e-04h 1
83 -1.6662900e+03 1.10e+04 9.14e+03 -2.5 3.34e+03 - 2.63e-04 2.01e-03h 1
84 -1.6662804e+03 1.10e+04 9.12e+03 -2.5 3.34e+03 - 1.71e-04 2.75e-03h 1
85 -1.6662717e+03 1.10e+04 9.09e+03 -2.5 3.33e+03 - 2.07e-04 3.15e-03h 1
86 -1.6662651e+03 1.09e+04 9.06e+03 -2.5 3.32e+03 - 1.96e-04 3.20e-03h 1
87 -1.6662648e+03 1.09e+04 9.06e+03 -2.5 3.31e+03 - 1.32e-04 1.89e-04h 1
88 -1.6662608e+03 1.09e+04 9.03e+03 -2.5 3.31e+03 - 8.58e-04 2.89e-03h 1
89 -1.6662585e+03 1.09e+04 9.00e+03 -2.5 3.31e+03 - 2.08e-04 3.30e-03h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
90 -1.6662579e+03 1.08e+04 8.99e+03 -2.5 3.30e+03 - 1.55e-04 1.88e-03h 1
91 -1.6662579e+03 1.08e+04 8.98e+03 -2.5 3.29e+03 - 8.90e-05 1.20e-03h 1
92 -1.6662579e+03 1.08e+04 8.97e+03 -2.5 3.29e+03 - 1.43e-04 4.14e-04h 1
93 -1.6662584e+03 1.08e+04 8.96e+03 -2.5 3.29e+03 - 2.35e-04 1.55e-03h 1
94 -1.6662598e+03 1.08e+04 8.94e+03 -2.5 3.29e+03 - 2.09e-04 2.30e-03h 1
95 -1.6662623e+03 1.08e+04 8.91e+03 -2.5 3.28e+03 - 1.69e-04 2.57e-03h 1
96 -1.6662661e+03 1.07e+04 8.89e+03 -2.5 3.27e+03 - 1.32e-04 2.70e-03h 1
97 -1.6662705e+03 1.07e+04 8.87e+03 -2.5 3.27e+03 - 9.71e-05 2.54e-03h 1
98 -1.6662748e+03 1.07e+04 8.85e+03 -2.5 3.26e+03 - 6.58e-05 2.08e-03h 1
99 -1.6662823e+03 1.06e+04 8.82e+03 -2.5 3.25e+03 - 2.97e-05 3.15e-03h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
100 -1.6662826e+03 1.06e+04 8.82e+03 -2.5 3.25e+03 - 1.10e-05 8.98e-05h 1
101 -1.6709214e+03 3.85e+03 5.09e+03 -2.5 3.25e+03 - 6.19e-05 1.00e+00f 1
102 -1.7047495e+03 1.89e+03 2.40e+03 -2.5 5.09e+02 - 1.47e-01 1.00e+00h 1
103 -1.7004577e+03 1.87e+03 2.38e+03 -2.5 9.17e+02 - 4.95e-01 7.93e-03h 1
104 -1.7042565e+03 1.82e+03 2.32e+03 -2.5 3.55e+02 - 4.50e-01 2.99e-02h 1
105 -1.8338727e+03 9.07e+02 1.15e+03 -2.5 3.54e+02 - 2.84e-01 1.00e+00h 1
106 -1.8728927e+03 6.99e+02 8.87e+02 -2.5 2.68e+02 - 9.59e-01 2.97e-01h 1
107 -1.9828660e+03 3.50e+02 4.43e+02 -2.5 2.24e+02 - 1.00e+00 1.00e+00h 1
108 -2.0025487e+03 1.98e+02 2.51e+02 -2.5 5.16e+01 - 6.87e-01 7.66e-01h 1
109 -2.0101950e+03 1.01e+02 1.27e+02 -2.5 5.07e+01 - 1.00e+00 9.72e-01h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
110 -2.0101958e+03 1.00e+02 1.27e+02 -2.5 1.00e+02 - 6.68e-01 1.39e-03h 1
111r-2.0101958e+03 1.00e+02 1.00e+03 2.0 0.00e+00 - 0.00e+00 4.91e-07R 4
112r-2.0100388e+03 1.00e+02 9.99e+02 2.0 5.94e+04 - 2.49e-03 4.76e-05f 1
113r-2.0025462e+03 1.03e+02 1.00e+03 0.6 2.80e+04 - 6.52e-05 3.20e-03f 1
114r-2.0095537e+03 1.05e+02 9.98e+02 0.6 2.71e+04 - 1.08e-03 1.81e-03f 1
115r-2.0154321e+03 1.07e+02 9.97e+02 0.6 1.82e+04 - 1.62e-04 1.99e-03f 1
116r-2.0195970e+03 1.08e+02 9.95e+02 0.6 1.35e+04 - 6.89e-04 1.68e-03f 1
117r-2.0237942e+03 1.09e+02 9.93e+02 0.6 1.24e+04 - 2.15e-03 1.92e-03f 1
118r-2.0298846e+03 1.09e+02 9.90e+02 0.6 9.82e+03 - 4.63e-03 2.96e-03f 1
119r-2.0384854e+03 1.11e+02 9.86e+02 0.6 7.05e+03 - 1.10e-02 4.51e-03f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
120r-2.0509912e+03 1.11e+02 5.12e+03 0.6 5.98e+03 - 4.14e-02 7.58e-03f 1
121r-2.0698248e+03 1.11e+02 8.12e+03 0.6 4.19e+03 - 4.22e-02 1.43e-02f 1
122r-2.0881157e+03 1.12e+02 1.82e+04 0.6 3.21e+03 - 1.41e-01 1.75e-02f 1
123r-2.1113751e+03 1.11e+02 2.18e+04 0.6 2.21e+03 - 1.21e-01 3.09e-02f 1
124r-2.1267149e+03 1.06e+02 3.00e+04 0.6 1.54e+03 - 3.62e-01 2.93e-02f 1
125r-2.1428597e+03 9.95e+01 2.92e+04 0.6 9.30e+02 - 3.40e-01 4.76e-02f 1
126r-2.1547936e+03 8.90e+01 2.54e+04 0.6 8.30e+02 - 4.58e-01 5.06e-02f 1
127 -2.1855501e+03 4.80e-01 1.05e+01 -2.5 6.97e+01 - 6.49e-01 9.97e-01h 1
128 -2.1853909e+03 4.87e-04 5.71e+01 -2.5 3.77e-01 - 9.25e-01 9.99e-01h 1
129 -2.1855496e+03 1.01e-04 1.99e+05 -2.5 2.63e-01 - 3.52e-02 1.00e+00f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
130 -2.3394033e+03 3.44e+04 2.63e+05 -2.5 5.04e+02 - 5.84e-04 5.06e-01f 1
131 -2.2534031e+03 2.22e+04 2.03e+05 -2.5 9.20e+02 - 1.93e-01 5.00e-01h 2
132 -2.2082517e+03 2.17e+04 1.89e+05 -2.5 9.62e+03 - 6.62e-02 2.43e-02h 6
133 -2.1854414e+03 2.15e+04 1.77e+05 -2.5 1.48e+04 - 5.94e-02 7.30e-03h 7
134 -2.1847277e+03 2.15e+04 1.67e+05 -2.5 1.54e+04 - 6.63e-02 2.12e-04h 12
135 -2.1843709e+03 2.15e+04 1.67e+05 -2.5 1.56e+04 - 7.09e-02 1.05e-04h 13
136 -7.2312935e+02 2.96e+04 3.53e+05 -2.5 1.52e+04 - 8.04e-02 4.38e-01h 1
137 -7.6288462e+02 1.28e+04 2.06e+05 -2.5 1.10e+03 -4.3 5.61e-02 1.00e+00h 1
138 -1.5794751e+03 1.80e+04 1.36e+05 -2.5 1.14e+04 -4.7 2.20e-01 1.00e+00h 1
139 -2.1381827e+03 1.05e+04 8.03e+04 -2.5 4.04e+03 -4.3 3.93e-01 1.00e+00h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
140 -2.4082459e+03 4.22e+03 4.60e+04 -2.5 2.09e+03 -3.9 6.57e-01 1.00e+00h 1
141 -2.6360795e+03 1.76e+03 2.35e+04 -2.5 8.97e+02 -3.5 9.02e-01 1.00e+00h 1
142 -2.7892466e+03 8.03e+02 1.15e+04 -2.5 4.77e+02 -3.0 9.25e-01 1.00e+00h 1
143 -2.7582316e+03 3.25e+02 5.66e+03 -2.5 1.34e+02 -2.6 9.85e-01 1.00e+00h 1
144 -2.6864739e+03 1.83e+02 2.50e+03 -2.5 1.78e+02 -3.1 1.00e+00 1.00e+00h 1
145 -2.6285441e+03 4.37e+01 8.50e+02 -2.5 1.03e+02 -2.7 1.00e+00 1.00e+00h 1
146 -2.6162486e+03 5.37e+00 1.36e+02 -2.5 1.88e+01 -3.1 1.00e+00 1.00e+00h 1
147 -2.6205639e+03 2.49e-02 1.80e+00 -2.5 8.15e+00 -3.6 1.00e+00 1.00e+00h 1
148 -2.8300964e+03 1.50e+03 4.49e+01 -2.5 3.82e+02 - 1.00e+00 1.00e+00f 1
149 -2.7758742e+03 6.40e+02 2.15e+01 -2.5 9.63e+01 - 1.00e+00 1.00e+00h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
150 -2.7264449e+03 1.68e+02 8.71e+00 -2.5 1.07e+02 - 1.00e+00 1.00e+00h 1
151 -2.7239427e+03 6.53e+00 1.29e+00 -2.5 5.22e+01 - 1.00e+00 1.00e+00h 1
152 -2.7238103e+03 1.71e-01 3.48e-02 -2.5 1.37e+01 - 1.00e+00 1.00e+00h 1
153 -2.7236895e+03 1.16e-04 2.36e-05 -2.5 1.94e-01 - 1.00e+00 1.00e+00h 1
154 -2.7236899e+03 5.79e-04 4.74e+01 -5.7 5.37e-01 - 1.00e+00 1.00e+00h 1
155 -2.7236897e+03 7.00e-10 1.85e-11 -5.7 3.17e-04 - 1.00e+00 1.00e+00h 1
156 -2.7236897e+03 2.47e-10 7.14e-13 -8.6 3.50e-04 - 1.00e+00 1.00e+00h 1
Number of Iterations....: 156
(scaled) (unscaled)
Objective...............: -2.7236897097349824e+03 -2.7236897097349824e+03
Dual infeasibility......: 7.1357223967104319e-13 7.1357223967104319e-13
Constraint violation....: 2.4692781153135002e-10 2.4692781153135002e-10
Complementarity.........: 2.5066260979429508e-09 2.5066260979429508e-09
Overall NLP error.......: 2.5066260979429508e-09 2.5066260979429508e-09
Number of objective function evaluations = 242
Number of objective gradient evaluations = 143
Number of equality constraint evaluations = 242
Number of inequality constraint evaluations = 0
Number of equality constraint Jacobian evaluations = 158
Number of inequality constraint Jacobian evaluations = 0
Number of Lagrangian Hessian evaluations = 156
Total CPU secs in IPOPT (w/o function evaluations) = 0.692
Total CPU secs in NLP function evaluations = 0.024
EXIT: Optimal Solution Found.
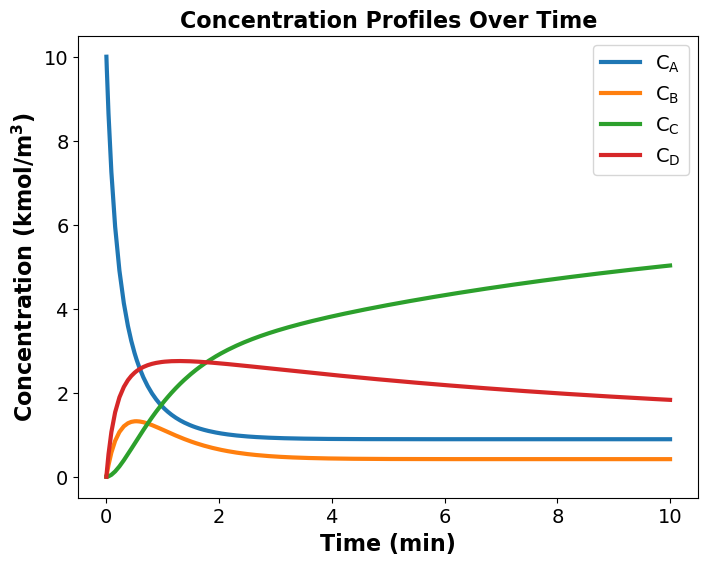
Optimal reactor volume: 9.02 m^3
Maximum Concentration of C at optimal reactor volume is: 5.03 kmol/m^3
Maximum Concentration of B at optimal reactor volume is: 0.42 kmol/m^3
# Visualizing Pareto Front
def generate_weights(n_points): # Function to systematically generate weight combinations
"""Creates weights for objective function
Argument:
n_points: number of points to evaluate weights
Returns:
w1: weight vector for objective 1
w2: weight vector for objective 2"""
w1 = np.linspace(0, 1, n_points)
w2 = [1 - w1[i] for i in range(len(w1))]
return w1, w2
# number of points for weights
n_points = 60
# Generate valid weight combinations
w1, w2 = generate_weights(n_points)
# Generate Pareto front by scanning weights
pareto_data = []
# Iterate over each weight combination
for i in range(len(w1)):
# Create and solve the model with the given weights
reactor_model = weighted_reactor_model(feed_flowrate, conc_A_feed, k1, k2, k3, w1[i], w2[i])
discretizer = pyo.TransformationFactory('dae.collocation')
discretizer.apply_to(reactor_model, nfe=20, ncp=10)
solver = pyo.SolverFactory('ipopt')
results = solver.solve(reactor_model, tee=False)
if results.solver.termination_condition == pyo.TerminationCondition.optimal:
optimal_volume = pyo.value(reactor_model.V)
optimal_CC = pyo.value(reactor_model.CC[10]) * 1e-3 # kmol/m^3
optimal_CB = pyo.value(reactor_model.CB[10]) * 1e-3 # kmol/m^3
pareto_data.append((w1[i], w2[i], optimal_CC, optimal_CB, optimal_volume))
# Convert Pareto data to a structured format for analysis
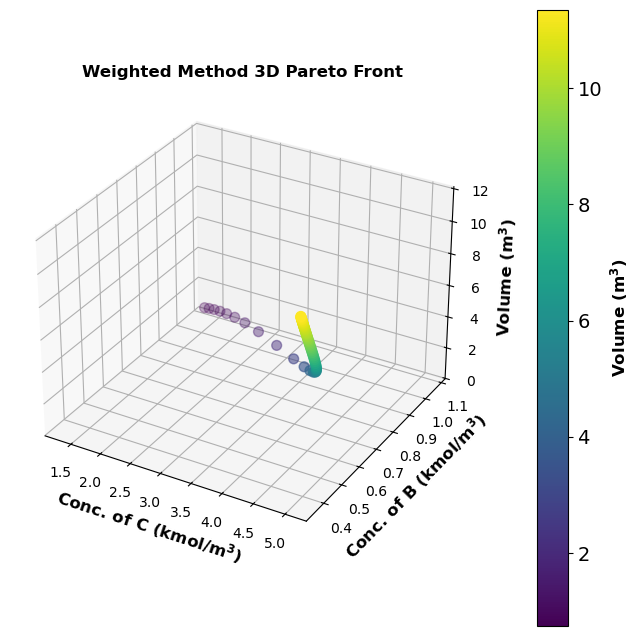
pareto_df = pd.DataFrame(pareto_data, columns=["w1", "w2", "CC", "CB", "V"])
# Plotting the 3D Pareto front
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(projection='3d')
scatter = ax.scatter(pareto_df['CC'], pareto_df['CB'], pareto_df['V'], c=pareto_df['V'], cmap='viridis', s=50)
ax.tick_params(axis='x', labelsize=10)
ax.tick_params(axis='y', labelsize=10)
ax.tick_params(axis='z', labelsize=10)
ax.tick_params(direction="in",top=True, right=True)
ax.set_xlabel('Conc. of C (kmol/$\mathbf{m^3}$)', fontsize=12,fontweight='bold')
ax.set_ylabel('Conc. of B (kmol/$\mathbf{m^3}$)', fontsize=12,fontweight='bold')
ax.set_zlabel('Volume ($\mathbf{m^3}$)', fontsize=12,fontweight='bold')
ax.set_title('Weighted Method 3D Pareto Front', fontsize=12,fontweight='bold')
color_bar = fig.colorbar(scatter, ax=ax, pad=0.1)
color_bar.set_label('Volume ($\mathbf{m^3}$)', fontsize=12,fontweight='bold')
plt.show()
Epsilon-Constraint Method Pyomo Implementation#
# Function for epsilon-constraint method multi-objective dynamic model
def create_epsilon_model(F, CAF, k1, k2, k3, epsilon_CB):
"""Solves multi-objective optimization using epsilon constraint method
Arguments:
F: feed volumetric flow rate in m^3/min
CAF: feed concentration of A, gmol/m^3
k1: rate constant for A to B, min^-1
k2: rate constant for B to C, min^-1
k3: rate constant for 2A to D, m^3/(gmol.min)
epsilon_CB: constraint threshold of concentration of B
Returns:
model: pyomo model
"""
model = pyo.ConcreteModel()
# Define time as a continuous set
model.t = dae.ContinuousSet(bounds=(0, 10))
# Variables
model.CA = pyo.Var(model.t, within=pyo.NonNegativeReals)
model.CB = pyo.Var(model.t, within=pyo.NonNegativeReals)
model.CC = pyo.Var(model.t, within=pyo.NonNegativeReals)
model.CD = pyo.Var(model.t, within=pyo.NonNegativeReals)
model.V = pyo.Var(within=pyo.NonNegativeReals, bounds=(0, 100), initialize=1)
# Derivatives
model.dCA_dt = dae.DerivativeVar(model.CA, wrt=model.t)
model.dCB_dt = dae.DerivativeVar(model.CB, wrt=model.t)
model.dCC_dt = dae.DerivativeVar(model.CC, wrt=model.t)
model.dCD_dt = dae.DerivativeVar(model.CD, wrt=model.t)
# CA differential equation
def ca_rate_ode(m, t):
return m.dCA_dt[t] == (F / m.V) * CAF - (F / m.V) * m.CA[t] - k1 * m.CA[t] - 2 * k3 * m.CA[t] ** 2
model.CA_ode = pyo.Constraint(model.t, rule=ca_rate_ode)
# CB differential equation
def cb_rate_ode(m, t):
return m.dCB_dt[t] == -(F / m.V) * m.CB[t] + k1 * m.CA[t] - k2 * m.CB[t]
model.CB_ode = pyo.Constraint(model.t, rule=cb_rate_ode)
# CC differential equation
def cc_rate_ode(m, t):
return m.dCC_dt[t] == -(F / m.V) * m.CC[t] + k2 * m.CB[t]
model.CC_ode = pyo.Constraint(model.t, rule=cc_rate_ode)
# CD differential equation
def cd_rate_ode(m, t):
return m.dCD_dt[t] == -(F / m.V) * m.CD[t] + k3 * m.CA[t] ** 2
model.CD_ode = pyo.Constraint(model.t, rule=cd_rate_ode)
# Initial conditions
def init_conditions(m):
yield m.CA[0] == CAF
yield m.CB[0] == 0
yield m.CC[0] == 0
yield m.CD[0] == 0
model.init_conditions = pyo.ConstraintList(rule=init_conditions)
# Epsilon constraints
model.epsilon_constraint_CB = pyo.Constraint(expr=model.CB[10] >= epsilon_CB) # converting CB to constraint
# Objective: Minimize volume
model.objective = pyo.Objective(expr=model.CC[10], sense=pyo.maximize)
return model
# constraint threshold
epsilon_CB_values = 420
# Create reactor model
reactor_model_epsilon = create_epsilon_model(feed_flowrate, conc_A_feed, k1, k2, k3, epsilon_CB_values)
# Discretize the model using collocation
discretizer = pyo.TransformationFactory('dae.collocation')
discretizer.apply_to(reactor_model_epsilon, nfe=20, ncp=10)
solver = pyo.SolverFactory('ipopt')
result = solver.solve(reactor_model_epsilon, tee=True)
if (result.solver.status == pyo.SolverStatus.ok) and (result.solver.termination_condition == pyo.TerminationCondition.optimal):
print("Solver found an optimal solution.")
optimal_volume = pyo.value(reactor_model_epsilon.V)
final_concentration_C = pyo.value(reactor_model_epsilon.CC[10]) * 1e-3
final_concentration_B = pyo.value(reactor_model_epsilon.CB[10]) * 1e-3
print(f"Optimal Reactor Volume: {optimal_volume:.2f} m^3")
print(f"Maximum Concentration of B at optimal reactor volume is: {final_concentration_B:.2f} kmol/m^3")
print(f"Maximum Concentration of C at optimal reactor volume is: {final_concentration_C:.2f} kmol/m^3")
else:
print("Solver did not find an optimal solution. Status:", result.solver.status)
# Extract results for plotting
time = [t for t in reactor_model_epsilon.t] # minutes
CA_values = [(pyo.value(reactor_model_epsilon.CA[t])*10**(-3)) for t in time] # kmol/m^3
CB_values = [(pyo.value(reactor_model_epsilon.CB[t])*10**(-3)) for t in time] # kmol/m^3
CC_values = [(pyo.value(reactor_model_epsilon.CC[t])*10**(-3)) for t in time] # kmol/m^3
CD_values = [(pyo.value(reactor_model_epsilon.CD[t])*10**(-3)) for t in time] # kmol/m^3
# Plot the results
plt.figure(figsize=(8, 6))
plt.plot(time, CA_values, label='$\mathrm{C_{A}}$')
plt.plot(time, CB_values, label='$\mathrm{C_{B}}$')
plt.plot(time, CC_values, label='$\mathrm{C_{C}}$')
plt.plot(time, CD_values, label='$\mathrm{C_{D}}$')
plt.xlabel('Time (min)', fontsize=16,fontweight='bold')
plt.ylabel('Concentration (kmol/$\mathbf{m^3}$)', fontsize=16,fontweight='bold')
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.title('Concentration Profiles Over Time', fontsize=16,fontweight='bold')
plt.legend()
plt.show()
Ipopt 3.13.2:
******************************************************************************
This program contains Ipopt, a library for large-scale nonlinear optimization.
Ipopt is released as open source code under the Eclipse Public License (EPL).
For more information visit http://projects.coin-or.org/Ipopt
This version of Ipopt was compiled from source code available at
https://github.com/IDAES/Ipopt as part of the Institute for the Design of
Advanced Energy Systems Process Systems Engineering Framework (IDAES PSE
Framework) Copyright (c) 2018-2019. See https://github.com/IDAES/idaes-pse.
This version of Ipopt was compiled using HSL, a collection of Fortran codes
for large-scale scientific computation. All technical papers, sales and
publicity material resulting from use of the HSL codes within IPOPT must
contain the following acknowledgement:
HSL, a collection of Fortran codes for large-scale scientific
computation. See http://www.hsl.rl.ac.uk.
******************************************************************************
This is Ipopt version 3.13.2, running with linear solver ma27.
Number of nonzeros in equality constraint Jacobian...: 12619
Number of nonzeros in inequality constraint Jacobian.: 1
Number of nonzeros in Lagrangian Hessian.............: 1006
Total number of variables............................: 1609
variables with only lower bounds: 804
variables with lower and upper bounds: 1
variables with only upper bounds: 0
Total number of equality constraints.................: 1608
Total number of inequality constraints...............: 1
inequality constraints with only lower bounds: 1
inequality constraints with lower and upper bounds: 0
inequality constraints with only upper bounds: 0
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
0 -9.9999900e-03 1.00e+04 2.28e+00 -1.0 0.00e+00 - 0.00e+00 0.00e+00 0
1 -4.8598389e-03 1.00e+04 3.18e+01 -1.0 1.89e+04 - 9.90e-07 3.28e-05h 1
2 -4.8257425e-03 1.00e+04 3.18e+01 -1.0 1.83e+04 - 3.34e-05 3.81e-05h 1
3 -9.4551719e-03 9.90e+03 3.90e+04 -1.0 1.83e+04 - 7.14e-05 9.73e-03f 1
4 -9.5086577e-03 9.90e+03 3.90e+04 -1.0 1.86e+04 - 2.53e-04 9.82e-05f 1
5r-9.5086577e-03 9.90e+03 9.99e+02 4.0 0.00e+00 - 0.00e+00 4.92e-07R 2
6r-1.1686406e+01 9.90e+03 8.24e+05 4.0 1.53e+04 - 8.22e-06 2.46e-03f 1
7r-1.6308831e+01 9.87e+03 4.90e+05 4.0 8.22e+02 - 5.17e-01 2.20e-01f 1
8r-1.3016905e+01 9.88e+03 3.58e+05 3.3 3.49e+02 - 5.65e-01 5.32e-01f 1
9r-1.3735757e+01 9.88e+03 2.13e+05 3.3 1.04e+02 - 1.00e+00 7.03e-01f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
10r-1.0871961e+01 9.89e+03 1.05e+05 3.3 2.81e+01 - 8.29e-01 1.00e+00f 1
11r-9.4210741e+00 9.91e+03 5.95e+04 2.6 4.06e+01 - 1.00e+00 7.51e-01f 1
12r-1.1429075e+01 9.91e+03 2.98e+04 2.6 1.25e+02 - 1.00e+00 8.86e-01f 1
13r-1.1915976e+01 9.91e+03 1.30e+04 2.6 1.29e+01 - 1.00e+00 1.00e+00f 1
14r-9.9938062e+00 9.93e+03 7.86e+03 1.9 5.17e+01 - 7.28e-01 6.37e-01f 1
15r-1.1032288e+01 9.92e+03 4.99e+03 1.9 2.46e+02 - 1.00e+00 4.95e-01f 1
16r-9.5439326e+00 9.91e+03 2.67e+03 1.2 1.11e+02 - 7.60e-01 8.46e-01f 1
17r-1.0596755e+01 9.89e+03 2.12e+03 1.2 5.88e+02 - 6.54e-01 2.35e-01f 1
18r-1.2101202e+01 9.87e+03 1.49e+03 1.2 4.69e+02 - 1.00e+00 3.58e-01f 1
19r-1.2133836e+01 9.87e+03 5.80e+02 1.2 3.03e+02 - 1.00e+00 1.21e-02f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
20r-1.3127762e+01 9.83e+03 2.63e+02 0.5 3.01e+02 - 8.75e-01 8.24e-01f 1
21r-1.7684525e+01 9.77e+03 1.91e+02 0.5 1.36e+03 - 2.67e-01 3.32e-01f 1
22r-2.0529528e+01 9.73e+03 2.09e+02 -0.2 8.99e+02 - 4.24e-01 3.23e-01f 1
23r-2.1186706e+01 9.72e+03 3.16e+02 -0.2 3.42e+03 - 2.63e-01 1.87e-02f 1
24r-3.0164095e+01 9.60e+03 2.07e+02 -0.2 3.34e+03 - 2.18e-01 2.59e-01f 1
25r-3.2513482e+01 9.57e+03 2.33e+02 -0.2 2.42e+03 - 5.43e-01 9.20e-02f 1
26r-3.8644532e+01 9.49e+03 2.41e+02 -0.2 2.18e+03 - 1.00e+00 2.66e-01f 1
27r-5.4811872e+01 9.28e+03 5.94e+00 -0.2 1.55e+03 - 1.00e+00 9.79e-01f 1
28r-5.4592211e+01 9.28e+03 2.53e-02 -0.2 3.06e+01 - 1.00e+00 1.00e+00h 1
29r-5.4474224e+01 9.28e+03 7.13e+01 -2.4 1.73e+00 - 9.94e-01 7.16e-01f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
30r-5.7067904e+01 9.25e+03 1.08e+02 -2.4 3.49e+04 - 4.36e-02 6.34e-03f 1
31r-6.0576269e+01 9.21e+03 1.42e+02 -2.4 3.45e+04 - 4.61e-02 8.63e-03f 1
32r-6.3326429e+01 9.18e+03 1.41e+02 -2.4 3.41e+04 - 6.88e-03 6.82e-03f 1
33r-6.7503485e+01 9.13e+03 1.47e+02 -2.4 3.37e+04 - 1.89e-02 1.04e-02f 1
34r-7.1226433e+01 9.09e+03 2.11e+02 -2.4 3.30e+04 - 4.90e-02 9.43e-03f 1
35r-9.8261702e+01 8.78e+03 1.53e+02 -2.4 3.25e+04 - 4.23e-02 6.95e-02f 1
36 -9.8265110e+01 8.78e+03 3.84e+00 -1.0 1.46e+04 - 1.12e-07 3.99e-07f 1
37 -9.8265544e+01 8.78e+03 7.64e+00 -1.0 1.41e+04 - 8.05e-08 3.28e-08f 1
38 -9.8268084e+01 8.78e+03 3.92e+00 -1.0 1.40e+04 - 1.01e-07 1.83e-07f 1
39 -2.3207517e+03 1.42e+04 2.93e+08 -1.0 1.40e+04 - 3.18e-07 1.60e-01f 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
40 -2.4052259e+03 1.23e+04 6.63e+08 -1.0 2.54e+04 - 1.72e-05 9.90e-01f 1
41 -2.4527209e+03 1.18e+04 6.38e+08 -1.0 1.30e+04 0.0 3.33e-01 3.99e-02h 1
42r-2.4527209e+03 1.18e+04 1.00e+03 3.9 0.00e+00 -0.5 0.00e+00 4.63e-07R 4
43r-2.3350121e+03 1.01e+04 4.48e+04 3.9 1.17e+05 - 7.55e-03 2.27e-02f 1
44 -2.4417906e+03 8.89e+03 1.44e+03 -1.0 1.25e+04 - 1.98e-01 1.16e-01f 1
45 -2.4447617e+03 8.86e+03 1.44e+03 -1.0 1.10e+04 - 1.08e-01 2.99e-03h 1
46 -3.4820052e+03 1.01e+03 9.36e+03 -1.0 1.09e+04 - 7.34e-02 9.90e-01f 1
47 -4.5367172e+03 3.39e+02 1.73e+03 -1.0 1.07e+03 - 3.69e-01 9.90e-01f 1
48 -4.9748649e+03 6.98e+01 1.02e+03 -1.0 4.81e+02 - 8.17e-01 9.91e-01f 1
49 -5.0264431e+03 4.22e+00 1.14e+04 -1.0 6.99e+01 - 9.81e-01 1.00e+00h 1
iter objective inf_pr inf_du lg(mu) ||d|| lg(rg) alpha_du alpha_pr ls
50 -5.0273073e+03 1.63e-02 3.07e+04 -1.0 1.41e+00 - 9.97e-01 1.00e+00h 1
51 -5.0272545e+03 2.47e-05 7.31e-05 -1.0 1.22e-01 - 1.00e+00 1.00e+00h 1
52 -5.0273606e+03 1.18e-04 6.11e+05 -5.7 2.57e-01 - 1.00e+00 9.39e-01f 1
53 -5.0273605e+03 1.02e-10 2.19e-10 -5.7 8.81e-05 - 1.00e+00 1.00e+00h 1
54 -5.0273605e+03 9.46e-11 2.45e-13 -8.6 4.20e-06 - 1.00e+00 1.00e+00h 1
Number of Iterations....: 54
(scaled) (unscaled)
Objective...............: -5.0273605021725707e+03 -5.0273605021725707e+03
Dual infeasibility......: 2.4547299726352162e-13 2.4547299726352162e-13
Constraint violation....: 6.8639442846867003e-11 9.4587448984384550e-11
Complementarity.........: 2.5059121858108535e-09 2.5059121858108535e-09
Overall NLP error.......: 2.5059121858108535e-09 2.5059121858108535e-09
Number of objective function evaluations = 61
Number of objective gradient evaluations = 26
Number of equality constraint evaluations = 61
Number of inequality constraint evaluations = 61
Number of equality constraint Jacobian evaluations = 57
Number of inequality constraint Jacobian evaluations = 57
Number of Lagrangian Hessian evaluations = 54
Total CPU secs in IPOPT (w/o function evaluations) = 0.168
Total CPU secs in NLP function evaluations = 0.007
EXIT: Optimal Solution Found.
Solver found an optimal solution.
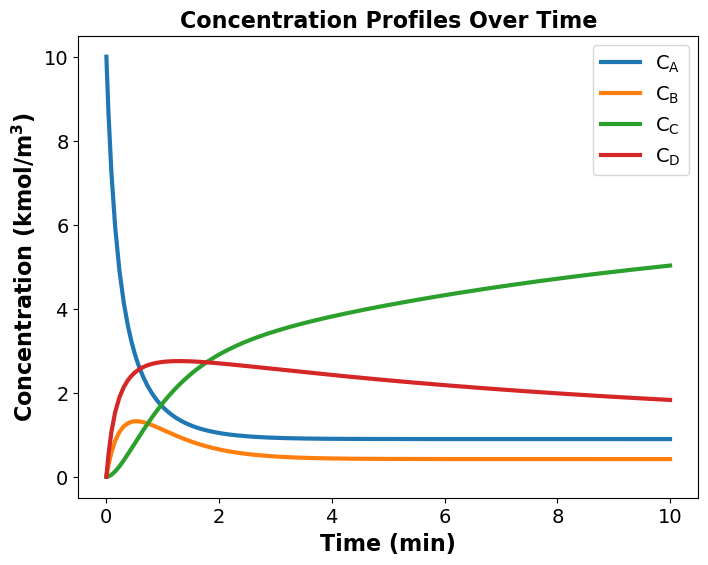
Optimal Reactor Volume: 8.97 m^3
Maximum Concentration of B at optimal reactor volume is: 0.42 kmol/m^3
Maximum Concentration of C at optimal reactor volume is: 5.03 kmol/m^3
# Visualizing Pareto Front
num_samples = 50 # sample amount
# constraint threshold
epsilon_CB_values = np.linspace(400, 700, num_samples)
# Store results
results = []
# loop through all constraint threshold values
for epsilon_CB in epsilon_CB_values:
reactor_model_epsilon = create_epsilon_model(feed_flowrate, conc_A_feed, k1, k2, k3, epsilon_CB) # reactor model
# Discretize the model using collocation
discretizer = pyo.TransformationFactory('dae.collocation')
discretizer.apply_to(reactor_model_epsilon, nfe=20, ncp=10)
solver = pyo.SolverFactory('ipopt')
result = solver.solve(reactor_model_epsilon, tee=False)
if (result.solver.status == pyo.SolverStatus.ok and result.solver.termination_condition == pyo.TerminationCondition.optimal):
optimal_volume = pyo.value(reactor_model_epsilon.V)
opt_concentration_CB = pyo.value(reactor_model_epsilon.CB[10])
opt_concentration_CC = pyo.value(reactor_model_epsilon.CC[10])
results.append({
'epsilon_CB': epsilon_CB,
'optimal_volume': optimal_volume,
'opt_concentration_CB': opt_concentration_CB,
'opt_concentration_CC': opt_concentration_CC
})
else:
print(f"Failed to find optimal solution for ε_CB = {epsilon_CB}")
results_df = pd.DataFrame(results)
print(results_df)
# Plotting the 3D Pareto front
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(projection='3d')
scatter = ax.scatter(results_df['opt_concentration_CC']*1e-3, results_df['opt_concentration_CB']*1e-3, results_df['optimal_volume'], c=results_df['optimal_volume'], cmap='viridis', s=50)
ax.tick_params(axis='x', labelsize=10)
ax.tick_params(axis='y', labelsize=10)
ax.tick_params(axis='z', labelsize=10)
ax.tick_params(direction="in",top=True, right=True)
ax.set_xlabel('Conc. of C (kmol/$\mathbf{m^3}$)', fontsize=12,fontweight='bold')
ax.set_ylabel('Conc. of B (kmol/$\mathbf{m^3}$)', fontsize=12,fontweight='bold')
ax.set_zlabel('Volume ($\mathbf{m^3}$)', fontsize=12,fontweight='bold')
ax.set_title('Epsilon-Constrainted Method 3D Pareto Front', fontsize=12,fontweight='bold')
color_bar = fig.colorbar(scatter, ax=ax, pad=0.1)
color_bar.set_label('Volume ($\mathbf{m^3}$)', fontsize=12,fontweight='bold')
plt.show()
epsilon_CB optimal_volume opt_concentration_CB opt_concentration_CC
0 400.000000 9.643572 399.999996 5044.649783
1 406.122449 9.431184 406.122445 5040.039755
2 412.244898 9.225387 412.244894 5034.826198
3 418.367347 9.025888 418.367343 5029.012010
4 424.489796 8.832410 424.489792 5022.600211
5 430.612245 8.644690 430.612241 5015.593948
6 436.734694 8.462483 436.734690 5007.996486
7 442.857143 8.285555 442.857138 4999.811213
8 448.979592 8.113687 448.979587 4991.041629
9 455.102041 7.946670 455.102036 4981.691353
10 461.224490 7.784306 461.224485 4971.764111
11 467.346939 7.626410 467.346934 4961.263742
12 473.469388 7.472804 473.469383 4950.194188
13 479.591837 7.323319 479.591832 4938.559499
14 485.714286 7.177797 485.714281 4926.363821
15 491.836735 7.036085 491.836730 4913.611402
16 497.959184 6.898039 497.959179 4900.306584
17 504.081633 6.763523 504.081628 4886.453801
18 510.204082 6.632405 510.204077 4872.057574
19 516.326531 6.504561 516.326525 4857.122514
20 522.448980 6.379872 522.448974 4841.653310
21 528.571429 6.258227 528.571423 4825.654734
22 534.693878 6.139515 534.693872 4809.131631
23 540.816327 6.023636 540.816321 4792.088918
24 546.938776 5.910489 546.938770 4774.531582
25 553.061224 5.799982 553.061219 4756.464673
26 559.183673 5.692023 559.183668 4737.893303
27 565.306122 5.586528 565.306117 4718.822637
28 571.428571 5.483414 571.428566 4699.257896
29 577.551020 5.382601 577.551015 4679.204348
30 583.673469 5.284015 583.673464 4658.667302
31 589.795918 5.187582 589.795912 4637.652110
32 595.918367 5.093234 595.918361 4616.164154
33 602.040816 5.000903 602.040810 4594.208848
34 608.163265 4.910525 608.163259 4571.791629
35 614.285714 4.822040 614.285708 4548.917952
36 620.408163 4.735387 620.408157 4525.593286
37 626.530612 4.650509 626.530606 4501.823108
38 632.653061 4.567353 632.653055 4477.612896
39 638.775510 4.485866 638.775504 4452.968125
40 644.897959 4.405996 644.897953 4427.894256
41 651.020408 4.327695 651.020402 4402.396738
42 657.142857 4.250915 657.142851 4376.480990
43 663.265306 4.175613 663.265299 4350.152404
44 669.387755 4.101743 669.387748 4323.416331
45 675.510204 4.029264 675.510197 4296.278076
46 681.632653 3.958134 681.632646 4268.742889
47 687.755102 3.888314 687.755095 4240.815958
48 693.877551 3.819767 693.877544 4212.502397
49 700.000000 3.752455 699.999993 4183.807240
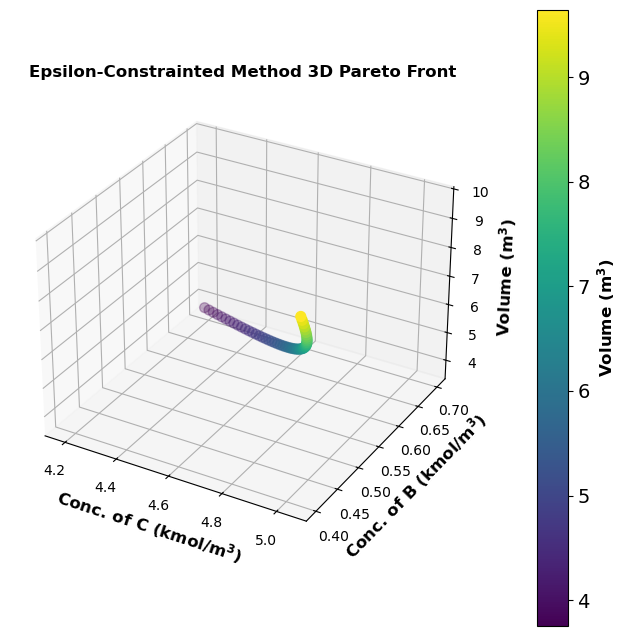
MOO Identifies the Lowest Reactor Volume and Offers Control over Production#
From the results of the two MOO methods, it can be seen that MOO identifies the lowest reactor volume (\(\mathrm{\sim 9\,m^3}\)) for an even higher species B concentration of \(\mathrm{0.42\, kmol/m^3}\), compared to single objective optimization in Extension 1 where the optimum reactor volume is \(\mathrm{11.4\,m^3}\) and species B concentration is \(\mathrm{0.36\,kmol/m^3}\).
MOO offer engineers the ability to select the product with highest priority, thereby enpowering stakeholders control over product yields.
Take Away Messages#
Pyomo continues to be a powerful tool in solving complex dynamic optimization problems.
Multi-objective optimization provides control over desired outcomes and products.
Multi-objective optimization identifies solutions that are robust to stakeholders decision.
Proper modeling of the problem is key to obtaining quality results.
Future work ideas:
Compare the Pareto sets generated with the two methods
Add another example with a non-convex Pareto set to highlight the difference between the methods
References#
Dowling, A. W., Ruiz-Mercado, G., & Zavala, V. M. (2016). A framework for multi-stakeholder decision-making and conflict resolution. Computers & Chemical Engineering, 90, 136-150.
Hart, W. E., Laird, C. D., Watson, J. P., Woodruff, D. L., Hackebeil, G. A., Nicholson, B. L., & Siirola, J. D. (2017). Pyomo-optimization modeling in python (Vol. 67, p. 277). Berlin: Springer.
Miettinen, K. (1999). Nonlinear multiobjective optimization (Vol. 12). Springer Science & Business Media.

