7.7. Simple Netwon Method for Equality Constrained NLPs#
Reference: Section 5.2 in Biegler (2010)

7.7.1. Helper Functions#
# Load required Python libraries.
import matplotlib.pyplot as plt
import numpy as np
from scipy import linalg
## Check is element of array is NaN
def check_nan(A):
return np.sum(np.isnan(A))
## Calculate gradient with central finite difference
## Calculate gradient with central finite difference
def my_grad_approx(x,f,eps1,verbose=False):
'''
Calculate gradient of function f using central difference formula
Inputs:
x - point for which to evaluate gradient
f - function to consider
eps1 - perturbation size
Outputs:
grad - gradient (vector)
'''
n = len(x)
grad = np.zeros(n)
if(verbose):
print("***** my_grad_approx at x = ",x,"*****")
for i in range(0,n):
# Create vector of zeros except eps in position i
e = np.zeros(n)
e[i] = eps1
# Finite difference formula
my_f_plus = f(x + e)
my_f_minus = f(x - e)
# Diagnostics
if(verbose):
print("e[",i,"] = ",e)
print("f(x + e[",i,"]) = ",my_f_plus)
print("f(x - e[",i,"]) = ",my_f_minus)
grad[i] = (my_f_plus - my_f_minus)/(2*eps1)
if(verbose):
print("***** Done. ***** \n")
return grad
def my_jac_approx(x,h,eps1,verbose=False):
'''
Calculate Jacobian of function h(x) using central difference formula
Inputs:
x - point for which to evaluate gradient
h - vector-valued function to consider. h(x): R^n --> R^m
eps1 - perturbation size
Outputs:
A - Jacobian (n x m matrix)
'''
# Check h(x) at x
h_x0 = h(x)
# Extract dimensions
n = len(x)
m = len(h_x0)
# Initialize Jacobian matrix
A = np.zeros((n,m))
# Calculate Jacobian by row
for i in range(0,n):
# Create vector of zeros except eps in position i
e = np.zeros(n)
e[i] = eps1
# Finite difference formula
my_h_plus = h(x + e)
my_h_minus = h(x - e)
# Diagnostics
if(verbose):
print("e[",i,"] = ",e)
print("h(x + e[",i,"]) = ",my_h_plus)
print("h(x - e[",i,"]) = ",my_h_minus)
A[i,:] = (my_h_plus - my_h_minus)/(2*eps1)
if(verbose):
print("***** Done. ***** \n")
return A
## Calculate gradient using central finite difference and my_hes_approx
def my_hes_approx(x,grad,eps2):
'''
Calculate gradient of function my_f using central difference formula and my_grad
Inputs:
x - point for which to evaluate gradient
grad - function to calculate the gradient
eps2 - perturbation size (for Hessian NOT gradient approximation)
Outputs:
H - Hessian (matrix)
'''
n = len(x)
H = np.zeros([n,n])
for i in range(0,n):
# Create vector of zeros except eps in position i
e = np.zeros(n)
e[i] = eps2
# Evaluate gradient twice
grad_plus = grad(x + e)
grad_minus = grad(x - e)
# Notice we are building the Hessian by column (or row)
H[:,i] = (grad_plus - grad_minus)/(2*eps2)
return H
## Linear algebra calculation
def xxT(u):
'''
Calculates u*u.T to circumvent limitation with SciPy
Arguments:
u - numpy 1D array
Returns:
u*u.T
Assume u is a nx1 vector.
Recall: NumPy does not distinguish between row or column vectors
u.dot(u) returns a scalar. This functon returns an nxn matrix.
'''
n = len(u)
A = np.zeros([n,n])
for i in range(0,n):
for j in range(0,n):
A[i,j] = u[i]*u[j]
return A
## Analyze Hessian
def analyze_hes(B):
print(B,"\n")
l = linalg.eigvals(B)
print("Eigenvalues: ",l,"\n")
7.7.2. Algorithm 5.1#
def alg51(x0,calc_f,calc_h,eps1=1E-6,eps2=1E-6,max_iter=50,verbose=False):
'''
Basic Full Space Newton Method for Equality Constrained NLP
Input:
x0 - starting point (vector)
calc_f - function to calculate objective (returns scalar)
calc_h - function to calculate constraints (returns vector)
eps1 - tolerance for primal and dual steps
eps2 - tolerance for gradient of L1
Outputs:
x - history of steps (primal variables)
v - history of steps (dual variables)
f - history of objective evaluations
h - history of constraint evaluations
df - history of objective gradients
dL - history of Lagrange function gradients
A - history of constraint Jacobians
W - history of Lagrange Hessians
Notes:
1. For simplicity, central finite difference is used
for all gradient calculations.
'''
# Declare iteration histories as empty lists
x = []
v = []
f = []
L = []
h = []
df = []
dL = []
A = []
W = []
# Flag for iterations
flag = True
# Iteration counter
k = 0
# Copy initial point to primal variable history
n = len(x0)
x.append(x0)
# Evaluate objective and constraints at initial point
f.append(calc_f(x0))
h.append(calc_h(x0))
# Determine number of equality constraints
m = len(h[0])
# Initial dual variables with vector of ones
v.append(np.ones(m))
# Print header for iteration information
print("Iter. \tf(x) \t\t||h(x)|| \t||grad_L(x)|| \t||dx|| \t\t||dv||")
while(flag and k < max_iter):
# STEP 1. Construct KKT matrix
if(k > 0):
# Evaluate objective function
f.append(calc_f(x[k]))
# Evaluate constraint function
h.append(calc_h(x[k]))
# Evaluate objective gradient
df.append(my_grad_approx(x[k],calc_f,1E-6))
# Evaluate constraint Jacobian
A.append(my_jac_approx(x[k],calc_h,1E-6))
# Evaluate gradient of Lagrange function
L_func = lambda x_ : calc_f(x_) + (calc_h(x_)).dot(v[k])
L_grad = lambda x_ : my_grad_approx(x_,L_func,1E-6)
dL.append(L_grad(x[k]))
norm_dL = linalg.norm(dL[k])
# Evaluate Hessian of Lagrange function
W.append(my_hes_approx(x[k],L_grad,1E-6))
if(verbose):
print("*** k =",k," ***")
print("x_k =",x[k])
print("v_k =",v[k])
print("f_k =",f[k])
print("df_k =",df[k])
print("h_k =",h[k])
print("A_k =\n",A[k])
print("W_k =\n",W[k])
print("\n")
# Assemble KKT matrix
KKT_top = np.concatenate((W[k],A[k]),axis=1)
KKT_bot = np.concatenate((A[k].T,np.zeros((m,m))),axis=1)
KKT = np.concatenate((KKT_top,KKT_bot),axis=0)
if(verbose):
print("KKT matrix =\n",KKT,"\n")
# Check if KKT matrix is singular
l, eigvec = linalg.eig(KKT)
if(verbose):
print("KKT matrix eigenvalues:")
print(l)
zero_eigenvalues = sum(np.abs(l) <= 1E-8)
if(zero_eigenvalues > 0):
flag = False
print("KKT matrix is singular. Eigenvalues:\n")
print(l,"\n")
## STEP 2. Solve linear system.
if(flag):
b = -np.concatenate((dL[k],h[k]),axis=0)
z = linalg.solve(KKT,b)
else:
z = []
## STEP 3. Take step
if(flag):
dx = z[0:n]
dv = z[n:(n+m+1)]
x.append(x[k] + dx)
v.append(v[k] + dv)
norm_dx = linalg.norm(dx)
norm_dv = linalg.norm(dv)
## Print iteration information
print(k,' \t{0: 1.4e} \t{1:1.4e} \t{2:1.4e}'.format(f[k],linalg.norm(h[k]),norm_dL),end='')
if flag:
print(' \t{0: 1.4e} \t{1: 1.4e}'.format(norm_dx,norm_dv),end='\n')
else:
print(' \t ------- \t -------',end='\n')
# Increment counter
k = k + 1
## Check convergence criteria
if(flag):
flag = norm_dx > eps1 and norm_dv > eps1 and norm_dL > eps2
if (not flag) and k >= max_iter:
print("Reached maximum number of iterations.")
return x,v,f,h,df,dL,A,W
7.7.3. Example Problem 1#
Consider:
\[\begin{split}\begin{align*} \min_{x} \quad & x_1^2 + 2 x_2^2 - x_3 \\
\mathrm{s.t.} \quad & x_1 + x_2 = 1 \\
& x_1 + x_2 - x_3 = 5
\end{align*} \end{split}\]
7.7.3.1. Define Functions#
def my_f(x):
return x[0]**2 + 2*x[1]**2 - 1*x[2]
def my_h(x):
h = np.zeros(2)
h[0] = x[0] + x[1] - 1
h[1] = x[0] + x[1] - x[2] - 5
return h
7.7.3.2. Test Finite Difference Approximations#
## Define initial point
x0 = np.array([1,1,1])
## Calculate objective
print("f(x0) =",my_f(x0),"\n")
## Calculate constraints
print("h(x0) =",my_h(x0),"\n")
## Calculate objective gradient
print("df(x0) =",my_grad_approx(x0,my_f,1E-6),"\n")
## Calculate constraint Jacobian
print("dh(x0) =\n",my_jac_approx(x0,my_h,1E-6),"\n")
f(x0) = 2
h(x0) = [ 1. -4.]
df(x0) = [ 2. 4. -1.]
dh(x0) =
[[ 1. 1.]
[ 1. 1.]
[ 0. -1.]]
7.7.3.3. Test Algorithm 5.1#
## Run Algorithm 5.1 on test problem
results = alg51(x0,my_f,my_h)
## Display results
xstar = results[0][-1]
print("\nx* =",xstar)
## Display results
vstar = results[1][-1]
print("\nv* =",vstar)
Iter. f(x) ||h(x)|| ||grad_L(x)|| ||dx|| ||dv||
0 2.0000e+00 4.1231e+00 7.4833e+00 5.0553e+00 2.4040e+00
1 4.6667e+00 5.7632e-10 8.2956e-04 1.5000e-04 6.2903e-04
2 4.6667e+00 0.0000e+00 1.4762e-08 1.9888e-09 8.1407e-09
x* = [ 0.66666667 0.33333333 -4. ]
v* = [-0.33333333 -1. ]
7.7.4. Example Problem 2#
Can we break Algorithm 5.1?
Probably. Let us try a model where \(\nabla h(x^k)^T\) is always rank-deficient.
Consider: $\(\begin{align}\min_x \quad & x_1^2 + 2 x_2^2 \\ \mathrm{s.t.} \quad & x_1 + x_2 = 1 \\ & x_1 + x_2 = 1 \end{align}\)$
7.7.4.1. Test Algorithm 5.1 with the redundant constraint#
def my_f2(x):
return x[0]**2 + 2*x[1]**2
def my_h2(x):
h = np.zeros(2)
h[0] = x[0] + x[1] - 1
h[1] = h[0]
return h
x0 = np.array((1,1))
## Run Algorithm 5.1 on test problem
results = alg51(x0,my_f2,my_h2)
## Display results
xstar = results[0][-1]
print("\nx* =",xstar)
## Display results
vstar = results[1][-1]
print("\nv* =",vstar)
Iter. f(x) ||h(x)|| ||grad_L(x)|| ||dx|| ||dv||
KKT matrix is singular. Eigenvalues:
[-1.05145076+0.j 2.51704031+0.j 4.53361158+0.j 0. +0.j]
0 3.0000e+00 1.4142e+00 7.2111e+00 ------- -------
x* = [1 1]
v* = [1. 1.]
7.7.4.2. Trying Algorithm 5.1 again without the redundant constraint#
def my_h2b(x):
return (x[0] + x[1] - 1)*np.ones(1)
x0 = np.array((1,1))
## Run Algorithm 5.1 on test problem
results = alg51(x0,my_f2,my_h2b)
## Display results
xstar = results[0][-1]
print("\nx* =",xstar)
## Display results
vstar = results[1][-1]
print("\nv* =",vstar)
Iter. f(x) ||h(x)|| ||grad_L(x)|| ||dx|| ||dv||
0 3.0000e+00 1.0000e+00 5.8310e+00 7.4537e-01 2.3335e+00
1 6.6667e-01 1.3978e-10 2.9473e-04 4.5326e-05 1.5285e-04
2 6.6667e-01 0.0000e+00 4.5431e-09 1.4393e-09 2.0170e-09
x* = [0.66666667 0.33333333]
v* = [-1.33333333]
7.7.5. Example Problem 3#
Can we break Algorithm 5.1 in another way?
Let us try a model where \(\nabla h(x^k)^T\) is full rank but there are multiple local optima.
Consider: $\(\begin{align}\min_x \quad & x_1^3 - x_2 -x_1 x_2 - x_2^2 \\ \mathrm{s.t.} \quad & x_1^2 + x_2^2 = 1 \end{align}\)$
def my_f3(x):
return x[0]**3 - x[1] - x[0]*x[1] - x[1]**2
def my_h3(x):
return (x[0]**2 + x[1]**2 - 1)*np.ones(1)
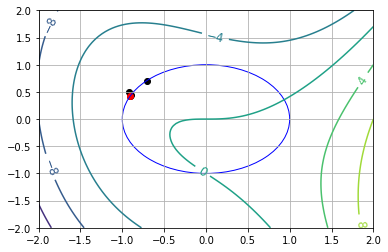
7.7.5.1. Visualize#
def visualize(xk=[]):
n1 = 101
n2 = 101
x1eval = np.linspace(-2,2,n1)
x2eval = np.linspace(-2,2,n2)
X, Y = np.meshgrid(x1eval, x2eval)
Z = np.zeros([n2,n1])
for i in range(0,n1):
for j in range(0,n2):
Z[j,i] = my_f3((X[j,i], Y[j,i]))
fig, ax = plt.subplots(1,1)
CS = ax.contour(X, Y, Z)
ax.clabel(CS, inline=1, fontsize=12)
# Add grid
plt.grid()
# Add unit circle
circ = plt.Circle((0, 0), radius=1, edgecolor='b', facecolor='None')
ax.add_patch(circ)
# Plot iteration history
if len(xk) > 0:
for i in range(0,len(xk)):
if i == len(xk) - 1:
c = "red"
else:
c = "black"
plt.scatter((xk[i][0]),(xk[i][1]),marker='o',color=c)
plt.xlim([-2,2])
plt.ylim([-2,2])
visualize()
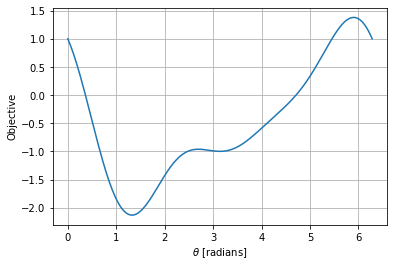
nt = 200
theta = np.linspace(0,2*np.pi,nt)
obj = np.zeros(nt)
for i in range(0,nt):
x_ = np.cos(theta[i])
y_ = np.sin(theta[i])
obj[i] = my_f3((x_,y_))
plt.figure()
plt.plot(theta,obj)
plt.xlabel('$\\theta$ [radians]')
plt.ylabel('Objective')
plt.grid()
7.7.5.2. Starting Point Near Global Min (\(\theta_0 = 1.0\))#
theta0 = 1.0
x0 = np.array((np.sin(theta0),np.cos(theta0)))
## Run Algorithm 5.1 on test problem
results = alg51(x0,my_f3,my_h3)
## Display results
xstar = results[0][-1]
print("\nx* =",xstar)
## Display results
vstar = results[1][-1]
print("\nv* =",vstar)
## Convert into theta
print("\ntheta* =",np.arccos(xstar[0]),"=",np.arcsin(xstar[1]))
## Visualize
visualize(results[0])
Iter. f(x) ||h(x)|| ||grad_L(x)|| ||dx|| ||dv||
0 -6.9105e-01 0.0000e+00 3.7501e+00 1.1171e+00 1.1455e+00
1 -4.0104e+00 1.2480e+00 2.1729e+00 4.2318e-01 3.9672e-01
2 -2.4216e+00 1.7908e-01 3.3575e-01 8.3649e-02 1.0205e-01
3 -2.1438e+00 6.9971e-03 1.7074e-02 3.5294e-03 6.5710e-03
4 -2.1324e+00 1.2457e-05 4.6327e-05 6.3597e-06 1.9707e-05
5 -2.1323e+00 4.0446e-11 1.4043e-09 2.8044e-10 3.3891e-11
x* = [0.24215301 0.97023807]
v* = [1.64012795]
theta* = 1.326212035697004 = 1.3262120356970035
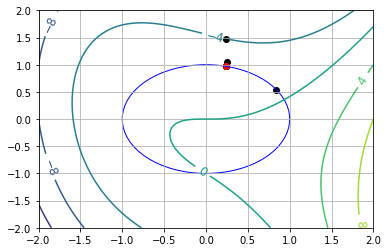
7.7.5.3. Starting Point Near Local Min (\(\theta_0 = \pi\))#
theta0 = np.pi
x0 = np.array((np.sin(theta0),np.cos(theta0)))
## Run Algorithm 5.1 on test problem
results = alg51(x0,my_f3,my_h3)
## Display results
xstar = results[0][-1]
print("\nx* =",xstar)
## Display results
vstar = results[1][-1]
print("\nv* =",vstar)
## Convert into theta
print("\ntheta* =",np.arccos(xstar[0]),"(using arccos)")
print("\ntheta* =",np.arcsin(xstar[1]),"(using arcsin)")
## Visualize
visualize(results[0])
Iter. f(x) ||h(x)|| ||grad_L(x)|| ||dx|| ||dv||
0 2.2204e-16 0.0000e+00 1.4142e+00 5.0002e-01 2.4998e-01
1 -6.2504e-01 2.5002e-01 1.0000e+00 1.5691e+00 5.7493e-01
2 1.4168e+00 2.4621e+00 4.6902e+00 8.7011e-01 3.2838e-01
3 -2.5316e-01 7.5709e-01 1.5056e+00 7.9530e-01 2.7969e-01
4 -1.0868e+00 6.3250e-01 1.3374e+00 8.5429e-01 2.8370e-01
5 -1.3530e-01 7.2981e-01 1.6069e+00 7.2946e-01 2.4685e-01
6 -8.5883e-01 5.3212e-01 1.1571e+00 1.2138e+00 4.1547e-01
7 6.1712e-01 1.4732e+00 3.1780e+00 7.4184e-01 2.5345e-01
8 -3.7711e-01 5.5033e-01 1.1886e+00 1.1024e+00 3.7605e-01
9 -2.5016e+00 1.2153e+00 2.6283e+00 7.1193e-01 2.4266e-01
10 -7.5418e-01 5.0685e-01 1.0966e+00 1.7975e+00 6.1304e-01
11 3.3538e+00 3.2310e+00 6.9879e+00 9.7525e-01 3.3253e-01
12 5.8930e-02 9.5111e-01 2.0572e+00 6.9821e-01 2.3817e-01
13 -6.1495e-01 4.8750e-01 1.0543e+00 1.4259e+01 4.8624e+00
14 2.5068e+03 2.0332e+02 4.3978e+02 7.1129e+00 1.9921e+00
15 3.1998e+02 5.0594e+01 1.0966e+02 3.5283e+00 1.0103e+00
16 4.2333e+01 1.2449e+01 2.7524e+01 1.7008e+00 4.4931e-02
17 7.8208e+00 2.8927e+00 6.7410e+00 7.3461e-01 8.5778e-02
18 2.3007e+00 5.3965e-01 1.6085e+00 2.1789e-01 2.8153e-01
19 1.4550e+00 4.7477e-02 2.3141e-01 2.3326e-02 8.3222e-02
20 1.3796e+00 5.4409e-04 5.3572e-03 2.8876e-04 2.4477e-03
21 1.3787e+00 8.3383e-08 1.4706e-06 1.0427e-07 5.7736e-07
x* = [ 0.92828695 -0.37186469]
v* = [-1.59272661]
theta* = 0.3810169551495443 (using arccos)
theta* = -0.3810169551495601 (using arcsin)
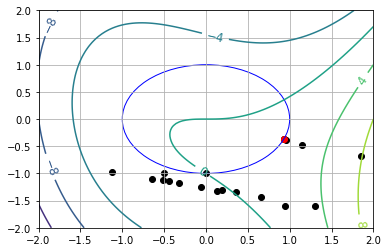
7.7.5.4. Starting Point Near Global Max (\(\theta_0 = 5.5\))#
theta0 = 5.5
x0 = np.array((np.sin(theta0),np.cos(theta0)))
## Run Algorithm 5.1 on test problem
results = alg51(x0,my_f3,my_h3)
## Display results
xstar = results[0][-1]
print("\nx* =",xstar)
## Display results
vstar = results[1][-1]
print("\nv* =",vstar)
## Convert into theta
print("\ntheta* =",np.arccos(xstar[0]),"(using arccos)")
print("\ntheta* =",np.arcsin(xstar[1]),"(using arcsin)")
## Visualize
visualize(results[0])
Iter. f(x) ||h(x)|| ||grad_L(x)|| ||dx|| ||dv||
0 -1.0621e+00 0.0000e+00 6.9215e-01 3.0715e-01 5.4165e-02
1 -1.0667e+00 9.4343e-02 1.2087e-01 5.8624e-02 5.2391e-02
2 -9.6641e-01 3.4368e-03 6.8422e-03 5.8274e-03 6.8831e-03
3 -9.6261e-01 3.3959e-05 8.0237e-05 9.3484e-05 8.7639e-05
4 -9.6258e-01 8.7393e-09 2.1977e-08 2.1636e-08 2.4039e-08
x* = [-0.90194813 0.43184437]
v* = [1.11352685]
theta* = 2.6950559970723194 (using arccos)
theta* = 0.4465366565174743 (using arcsin)